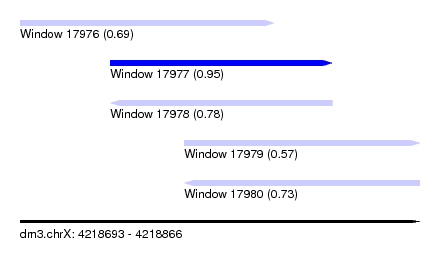
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,218,693 – 4,218,866 |
| Length | 173 |
| Max. P | 0.949005 |

| Location | 4,218,693 – 4,218,803 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 93.51 |
| Shannon entropy | 0.11207 |
| G+C content | 0.42196 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -21.70 |
| Energy contribution | -22.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.690540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4218693 110 + 22422827 GAUUGUGCAAUAAAGCUAUAAUAAUAAAAUAACAAACAAACCGUUAAAGCGAACGCCGUCGCACACUCAUCGCACCCACACAAACGCACCAUAGGUAUUGGUGUGUGGCU ((.(((((......(((.((((....................)))).)))((......))))))).))...((..........((((((((.......)))))))).)). ( -23.05, z-score = -0.56, R) >droSim1.chrX 3246765 110 + 17042790 GAUUGUGCAAUAAAGCUAUAAUAAUAAAAUAACAAACAAACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCAAAGGUAUAGGUGUGUUGGU ((.(((((.....................((((.........)))).(((....)))...))))).)).....(((((((((.....(((...))).....))))))))) ( -28.60, z-score = -2.53, R) >droSec1.super_4 3671092 110 - 6179234 GAUUGUGCAAUAAAGCUAUAAUAAUAAAAUAACAAACAAACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCAAAGGUAUAGGUGUGUUGGU ((.(((((.....................((((.........)))).(((....)))...))))).)).....(((((((((.....(((...))).....))))))))) ( -28.60, z-score = -2.53, R) >droYak2.chrX 17547589 104 - 21770863 GAUUGUGCAAUAAAGCUAUAAUAAUAAAAUAACAAACAAACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCA------UAGGUGUGUUGUU ((.(((((.....................((((.........)))).(((....)))...))))).))..............((((((((.------..))))))))... ( -24.10, z-score = -1.94, R) >droEre2.scaffold_4690 1588930 104 + 18748788 GAUUGUGCAAUAAAGCUAUAAUAAUAAAAUAACAAACAAACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAGCUCACCA------UAGGUGUGUGGGU ....((((.....................((((.........)))).(((....)))...))))(((((.((((((...............------..))))))))))) ( -23.93, z-score = -1.39, R) >consensus GAUUGUGCAAUAAAGCUAUAAUAAUAAAAUAACAAACAAACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCA_AGGUAUAGGUGUGUUGGU ((.(((((.....................((((.........)))).(((....)))...))))).))..............((((((((.........))))))))... (-21.70 = -22.34 + 0.64)

| Location | 4,218,732 – 4,218,828 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.78 |
| Shannon entropy | 0.28340 |
| G+C content | 0.55939 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -24.70 |
| Energy contribution | -26.94 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4218732 96 + 22422827 ACCGUUAAAGCGAACGCCGUCGCACACUCAUCGCACCCACACAAACGCACCAUAGGUAUUGGUGUGUGGCUGCAUGAUGGGGCGGAAGAAAUGGAG-- .((((....((((......))))...(((((((((.(((((((.....(((...))).....))))))).)))..))))))))))...........-- ( -33.80, z-score = -1.56, R) >droSim1.chrX 3246804 98 + 17042790 ACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCAAAGGUAUAGGUGUGUUGGUGCAUGAGGGGGUGGAAGAUAUAGGGAG .((.....(((....)))(((.(((.(((((.(((((((((((.....(((...))).....))))))))))))))))...)))...)))...))... ( -39.50, z-score = -4.02, R) >droSec1.super_4 3671131 98 - 6179234 ACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCAAAGGUAUAGGUGUGUUGGUGCAUGAGGGGGUGGAAGAUAUAGGGAG .((.....(((....)))(((.(((.(((((.(((((((((((.....(((...))).....))))))))))))))))...)))...)))...))... ( -39.50, z-score = -4.02, R) >droYak2.chrX 17547628 87 - 21770863 ACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCA------UAGGUGUGUUGUUGCACGAGGGG-UGGUGGAAAGAG---- ........(((....))).((((.(((((.(((...(((((...(((((((.------..))))))))))))..))).)))-))))))......---- ( -30.80, z-score = -1.36, R) >droEre2.scaffold_4690 1588969 90 + 18748788 ACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAGCUCACCA------UAGGUGUGUGGGUGCACGAUGCGGGGGAAGCGGAAGAG-- .(((((..(((....))).((.(...(.(((((((((.(((((..((.....------.)).))))).)))))..)))).).).))))))).....-- ( -31.80, z-score = -1.07, R) >consensus ACCGUUAAGGCGAACGCCGUCGCACACUCAUCGCACCAACACAAACGCACCA_AGGUAUAGGUGUGUUGGUGCAUGAGGGGGUGGAAGAAAUAGAG__ .(((((...((((......))))...(((((.(((((((((((.....((.....)).....))))))))))))))))..)))))............. (-24.70 = -26.94 + 2.24)
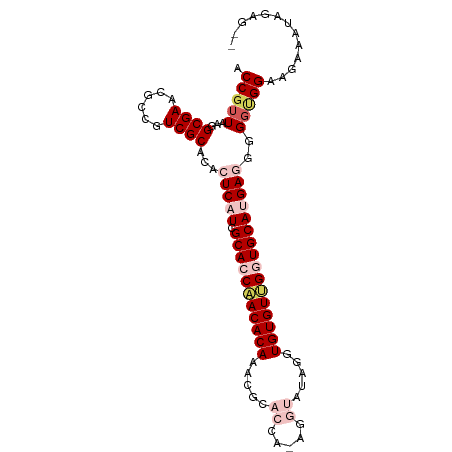
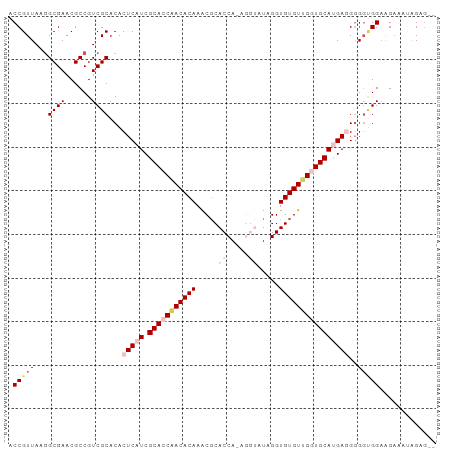
| Location | 4,218,732 – 4,218,828 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.78 |
| Shannon entropy | 0.28340 |
| G+C content | 0.55939 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -24.66 |
| Energy contribution | -25.02 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4218732 96 - 22422827 --CUCCAUUUCUUCCGCCCCAUCAUGCAGCCACACACCAAUACCUAUGGUGCGUUUGUGUGGGUGCGAUGAGUGUGCGACGGCGUUCGCUUUAACGGU --...........(((...((((..(((.(((((((....((((...))))....))))))).)))))))((((.(((....))).))))....))). ( -30.00, z-score = -0.39, R) >droSim1.chrX 3246804 98 - 17042790 CUCCCUAUAUCUUCCACCCCCUCAUGCACCAACACACCUAUACCUUUGGUGCGUUUGUGUUGGUGCGAUGAGUGUGCGACGGCGUUCGCCUUAACGGU ...((.......(((((...((((((((((((((((....((((...))))....))))))))))).))))).))).)).(((....))).....)). ( -35.10, z-score = -3.74, R) >droSec1.super_4 3671131 98 + 6179234 CUCCCUAUAUCUUCCACCCCCUCAUGCACCAACACACCUAUACCUUUGGUGCGUUUGUGUUGGUGCGAUGAGUGUGCGACGGCGUUCGCCUUAACGGU ...((.......(((((...((((((((((((((((....((((...))))....))))))))))).))))).))).)).(((....))).....)). ( -35.10, z-score = -3.74, R) >droYak2.chrX 17547628 87 + 21770863 ----CUCUUUCCACCA-CCCCUCGUGCAACAACACACCUA------UGGUGCGUUUGUGUUGGUGCGAUGAGUGUGCGACGGCGUUCGCCUUAACGGU ----......((.(((-(..((((((((.(((((((...(------((...))).))))))).))).))))).))).)..(((....))).....)). ( -24.50, z-score = 0.44, R) >droEre2.scaffold_4690 1588969 90 - 18748788 --CUCUUCCGCUUCCCCCGCAUCGUGCACCCACACACCUA------UGGUGAGCUUGUGUUGGUGCGAUGAGUGUGCGACGGCGUUCGCCUUAACGGU --.....(((.......(((((((((((((.(((((.((.------.....))..))))).))))).))))...))))..(((....)))....))). ( -28.70, z-score = -0.26, R) >consensus __CCCUAUAUCUUCCACCCCCUCAUGCACCAACACACCUAUACCU_UGGUGCGUUUGUGUUGGUGCGAUGAGUGUGCGACGGCGUUCGCCUUAACGGU .............((.....((((((((((((((((((.........)))......)))))))))).)))))........(((....))).....)). (-24.66 = -25.02 + 0.36)
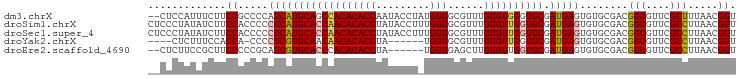

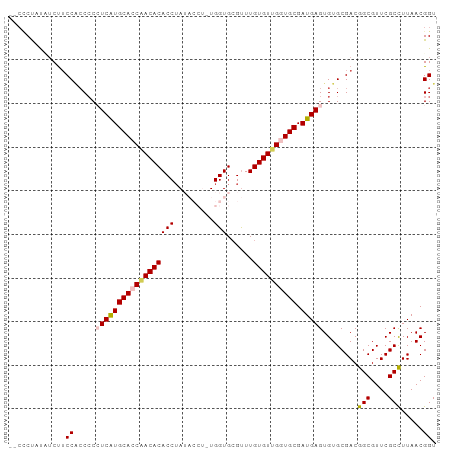
| Location | 4,218,764 – 4,218,866 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.07 |
| Shannon entropy | 0.32737 |
| G+C content | 0.50381 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -18.74 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
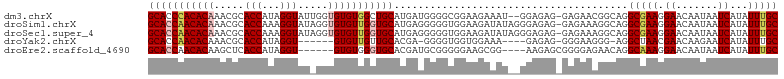
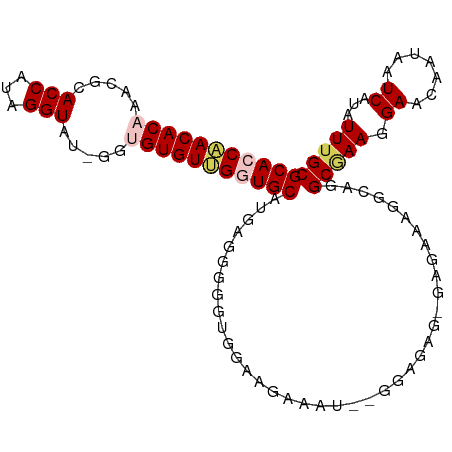
>dm3.chrX 4218764 102 + 22422827 GCACCCACACAAACGCACCAUAGGUAUUGGUGUGUGGCUGCAUGAUGGGGCGGAAGAAAU--GGAGAG-GAGAACGGCAGGCGAAGGAACAAUAAUCAUAUUUGC ((.((((((((.((((((((.......))))))))...))..)).)))).(....)....--......-.......))..(((((.((.......))...))))) ( -23.50, z-score = 0.03, R) >droSim1.chrX 3246836 104 + 17042790 GCACCAACACAAACGCACCAAAGGUAUAGGUGUGUUGGUGCAUGAGGGGGUGGAAGAUAUAGGGAGAG-GAGAAAGGCAGGCGAAGGAACAAUAAUCAUAUUUGC (((((((((((.....(((...))).....)))))))))))...........................-...........(((((.((.......))...))))) ( -23.90, z-score = -2.50, R) >droSec1.super_4 3671163 104 - 6179234 GCACCAACACAAACGCACCAAAGGUAUAGGUGUGUUGGUGCAUGAGGGGGUGGAAGAUAUAGGGAGAG-GAGAAAGGCAGGCGAAGGAACAAUAAUCAUAUUUGC (((((((((((.....(((...))).....)))))))))))...........................-...........(((((.((.......))...))))) ( -23.90, z-score = -2.50, R) >droYak2.chrX 17547660 92 - 21770863 GCACCAACACAAACGCACCAUAGGU------GUGUUGUUGCACGA-GGGGUGGUGGAAA----GAGAG-GGGAAGGG-AGGCUAACGAACAAGAAUCAUAUUUGC (((((..........((((((...(------((((....))))).-...))))))....----.....-.))....(-(..((........))..)).....))) ( -18.05, z-score = 0.52, R) >droEre2.scaffold_4690 1589001 95 + 18748788 GCACCAACACAAGCUCACCAUAGGU------GUGUGGGUGCACGAUGCGGGGGAAGCGG----AAGAGCGGGGAGAACAGGCAAAGGAACAAUAAUCAUAUUUGC (((((.(((((..((......)).)------)))).))))).....(((((.((..(..----..).((..(.....)..)).............))...))))) ( -21.30, z-score = -0.03, R) >consensus GCACCAACACAAACGCACCAUAGGUAU_GGUGUGUUGGUGCAUGAGGGGGUGGAAGAAAU__GGAGAG_GAGAAAGGCAGGCGAAGGAACAAUAAUCAUAUUUGC (((((((((((.....(((...))).....))))))))))).......................................(((((.((.......))...))))) (-18.74 = -19.58 + 0.84)
| Location | 4,218,764 – 4,218,866 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.07 |
| Shannon entropy | 0.32737 |
| G+C content | 0.50381 |
| Mean single sequence MFE | -17.54 |
| Consensus MFE | -14.82 |
| Energy contribution | -15.98 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
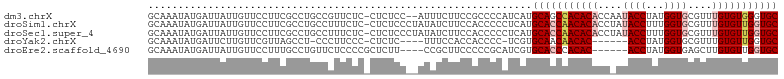
>dm3.chrX 4218764 102 - 22422827 GCAAAUAUGAUUAUUGUUCCUUCGCCUGCCGUUCUC-CUCUCC--AUUUCUUCCGCCCCAUCAUGCAGCCACACACCAAUACCUAUGGUGCGUUUGUGUGGGUGC (((....(((...........)))..))).......-......--...................(((.(((((((....((((...))))....))))))).))) ( -17.80, z-score = 0.51, R) >droSim1.chrX 3246836 104 - 17042790 GCAAAUAUGAUUAUUGUUCCUUCGCCUGCCUUUCUC-CUCUCCCUAUAUCUUCCACCCCCUCAUGCACCAACACACCUAUACCUUUGGUGCGUUUGUGUUGGUGC (((....(((...........)))..))).......-...........................(((((((((((....((((...))))....))))))))))) ( -19.90, z-score = -3.25, R) >droSec1.super_4 3671163 104 + 6179234 GCAAAUAUGAUUAUUGUUCCUUCGCCUGCCUUUCUC-CUCUCCCUAUAUCUUCCACCCCCUCAUGCACCAACACACCUAUACCUUUGGUGCGUUUGUGUUGGUGC (((....(((...........)))..))).......-...........................(((((((((((....((((...))))....))))))))))) ( -19.90, z-score = -3.25, R) >droYak2.chrX 17547660 92 + 21770863 GCAAAUAUGAUUCUUGUUCGUUAGCCU-CCCUUCCC-CUCUC----UUUCCACCACCCC-UCGUGCAACAACAC------ACCUAUGGUGCGUUUGUGUUGGUGC ((....((((.......))))..))..-........-.....----....(((((....-..(..(((..((.(------(((...)))).)))))..)))))). ( -13.80, z-score = 0.02, R) >droEre2.scaffold_4690 1589001 95 - 18748788 GCAAAUAUGAUUAUUGUUCCUUUGCCUGUUCUCCCCGCUCUU----CCGCUUCCCCCGCAUCGUGCACCCACAC------ACCUAUGGUGAGCUUGUGUUGGUGC (((((...((.......)).)))))...........((....----..)).......((((((.((((...(.(------(((...)))).)...)))))))))) ( -16.30, z-score = 0.24, R) >consensus GCAAAUAUGAUUAUUGUUCCUUCGCCUGCCCUUCUC_CUCUCC__AUAUCUUCCACCCCAUCAUGCACCAACACACC_AUACCUAUGGUGCGUUUGUGUUGGUGC ................................................................(((((((((((....((((...))))....))))))))))) (-14.82 = -15.98 + 1.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:15 2011