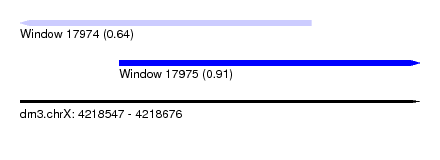
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,218,547 – 4,218,676 |
| Length | 129 |
| Max. P | 0.909999 |

| Location | 4,218,547 – 4,218,641 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.57 |
| Shannon entropy | 0.07105 |
| G+C content | 0.40957 |
| Mean single sequence MFE | -18.38 |
| Consensus MFE | -16.90 |
| Energy contribution | -16.65 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
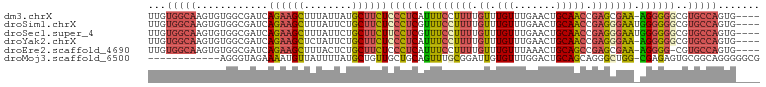
>dm3.chrX 4218547 94 - 22422827 AACAAACAAAAGGAAAUGAGGGAGAAGCAUAAUAAAGCUUCUGAUCGCCACACUUGCCACAAAAAGUAAGCUACGUAGAACUACAGUAGAGCUU ...........((((.((.(((((((((........)))))...)).)).)).)).)).....((((...(((((((....))).)))).)))) ( -17.20, z-score = -0.94, R) >droSim1.chrX 3246618 94 - 17042790 AACAAACAAAAGGAAACGAGGGAGAAGCAGAAUAAAGCUUCUGAUCGCCACACUUGCCACAAAAAGUAAGCUACGUAGAACUACAGUAGAGCUU ...........(....)((...((((((........))))))..))((....(((((........)))))(((((((....))).)))).)).. ( -19.00, z-score = -1.58, R) >droSec1.super_4 3670945 94 + 6179234 AACAAACAAAAGGAAACGAGGAAGAAGCAGAAUAAAGCUUCUGAUCGCCACACUUGCCACAAAAAGUAAGCUACGUAGAACUACAGUAGAGCUU ...........(....)((...((((((........))))))..))((....(((((........)))))(((((((....))).)))).)).. ( -20.10, z-score = -2.30, R) >droYak2.chrX 17547443 94 + 21770863 AACAAACAAAAGGAAAUGAGGGAGAAGCAGAAUAGAGCUUCUGAUCGCCACACUUGCCACAAAAAGUACCCUACGUAGUACUACAGUAGAGCAU ..................((((((((((........)))))).........((((........)))).))))..((..(((....)))..)).. ( -17.20, z-score = -0.90, R) >consensus AACAAACAAAAGGAAACGAGGGAGAAGCAGAAUAAAGCUUCUGAUCGCCACACUUGCCACAAAAAGUAAGCUACGUAGAACUACAGUAGAGCUU ...........((...(((...((((((........))))))..)))))..((((........))))...(((((((....))).))))..... (-16.90 = -16.65 + -0.25)
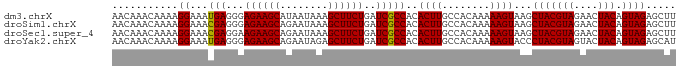
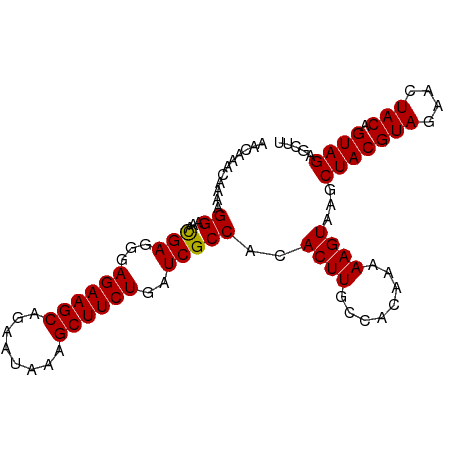
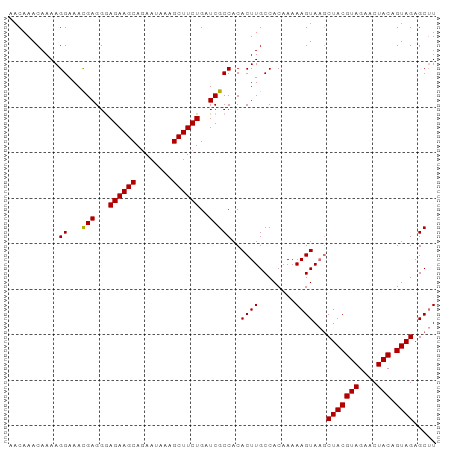
| Location | 4,218,579 – 4,218,676 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Shannon entropy | 0.42242 |
| G+C content | 0.50654 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
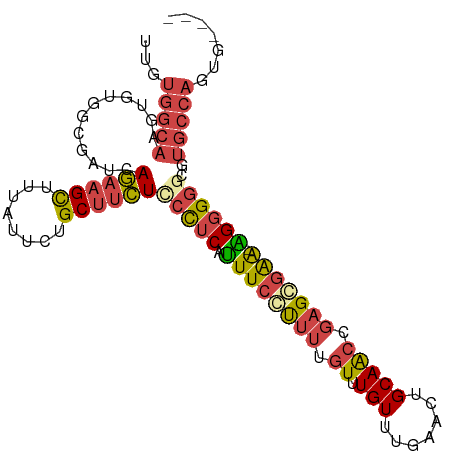
>dm3.chrX 4218579 97 + 22422827 UUGUGGCAAGUGUGGCGAUCAGAAGCUUUAUUAUGCUUCUCCCUCAUUUCCUUUUGUUUGUUUGAACUGCAACCGAGCGAA-AGGGGGCGUGCCAGUG---- ...(((((..((.((.((...(((((........))))))))).))(..(((((((((((.(((.....))).))))))))-)))..)..)))))...---- ( -33.80, z-score = -2.35, R) >droSim1.chrX 3246650 98 + 17042790 UUGUGGCAAGUGUGGCGAUCAGAAGCUUUAUUCUGCUUCUCCCUCGUUUCCUUUUGUUUGUUUGAACUGCAACCGAGGGAAUGGGGGGCGUGCCAGUG---- ...(((((.((.........((((((........))))))(((((((((((((((((..((....)).))))..))))))))))))))).)))))...---- ( -37.40, z-score = -2.72, R) >droSec1.super_4 3670977 98 - 6179234 UUGUGGCAAGUGUGGCGAUCAGAAGCUUUAUUCUGCUUCUUCCUCGUUUCCUUUUGUUUGUUUGAACUGCAACCGAGGGAAUGGGGGGCGUGCCAGUG---- ...(((((.((.........((((((........))))))(((((((((((((((((..((....)).))))..))))))))))))))).)))))...---- ( -34.70, z-score = -2.15, R) >droYak2.chrX 17547475 97 - 21770863 UUGUGGCAAGUGUGGCGAUCAGAAGCUCUAUUCUGCUUCUCCCUCAUUUCCUUUUGUUUGUUUGAACUGCAACCGAGGGAA-AGGGGGCGUGCCAGUG---- ...(((((............((((((........))))))(((((.((((((((.(((.((.......))))).)))))))-))))))..)))))...---- ( -32.70, z-score = -1.39, R) >droEre2.scaffold_4690 1588810 96 + 18748788 UUGUGGCAAGUGUGGCGAUCAGAAGCUUUACUCUGCUUCUCCCUCAUUUCCUUUUGUUUGUUUAAACUGCAGCCGAGCGAA-AGGGG-CGUGCCAGUG---- ...(((((((((.((.((...(((((........))))))))).))))((((((((((((.............))))))))-)))).-..)))))...---- ( -30.22, z-score = -1.03, R) >droMoj3.scaffold_6500 3100879 89 + 32352404 ------------AGGGUAGAAAAUGUUAUUUUAUGCUGUUGCUGCAGUUUGCGGAUUGUGUUUGGACUGCAGCAGGGCUGG-CGAGAGUGCGGCAGGGGGCG ------------...((((((.......))))))(((.(((((((((((((((.....)))..)))))))))))).(((((-(....)).))))....))). ( -28.30, z-score = -1.18, R) >consensus UUGUGGCAAGUGUGGCGAUCAGAAGCUUUAUUCUGCUUCUCCCUCAUUUCCUUUUGUUUGUUUGAACUGCAACCGAGCGAA_AGGGGGCGUGCCAGUG____ ...(((((............((((((........))))))((((...(((((((.((.(((.......))))).))))))).))))....)))))....... (-20.46 = -21.05 + 0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:12 2011