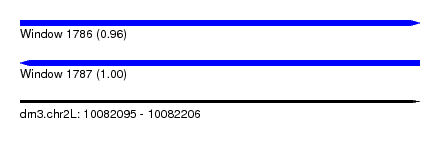
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 10,082,095 – 10,082,206 |
| Length | 111 |
| Max. P | 0.996127 |

| Location | 10,082,095 – 10,082,206 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 65.10 |
| Shannon entropy | 0.62417 |
| G+C content | 0.34690 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -10.82 |
| Energy contribution | -10.72 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.964288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
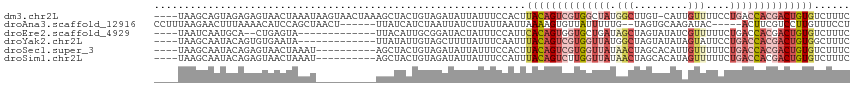
>dm3.chr2L 10082095 111 + 23011544 GAAAGACACAGUCGUGGUCAGGAAAACAAUG-ACAAGCCAUAGCCACGACUGUAAGUGGAAAUAAUAUCUACAGUAGCUUUAGUUACUUAUUUAGUUACUCUCUACUGCUUA---- ..(((.((((((((((((..((.........-.....))...))))))))))).((((((..((((......((((((....))))))......))))...)))))))))).---- ( -29.84, z-score = -2.47, R) >droAna3.scaffold_12916 13913125 103 + 16180835 AGGAAACAAGGACGAAGU-----GUAUCUUGCACUA--CAAAAAUAACACUUUUAAUUAAUAAGAUAAUUAGAUGAUAA------AGUUAGCUGGAUGUUUUAAAGUUCUUAAAGG .(....)(((((((.(((-----((.....))))).--).....................(((((((.((((.((((..------.)))).)))).)))))))..))))))..... ( -17.60, z-score = -1.24, R) >droEre2.scaffold_4929 10697908 97 + 26641161 GAAAGACACAGUCGUGGUCAGAAAAACGAUAUACUAGCUAUCAGCACCACUGUGAAUGGAAAUAGUAUCCGCAAUGUAA-------------UACUCAG--UGCAUUGAUUA---- ......((((((.(((((.......))((((.......))))..))).))))))...(((.......))).(((((((.-------------.......--)))))))....---- ( -19.30, z-score = -0.33, R) >droYak2.chr2L 12766175 99 + 22324452 GAAAGCCACAGUCGUGGUCAGGAAUACUAUAUACUAGCCAUAACCACGACUGUAAAUUGAAAUAAAAGCUACAAUAUAA-------------UAUUCACACUGUAUUGCUUA---- ..((((.(((((((((((..((....(((.....)))))...)))))))))))...................((((((.-------------.........)))))))))).---- ( -21.80, z-score = -2.72, R) >droSec1.super_3 5520304 102 + 7220098 GAAAGACACAGUCGUGGUCAGAAAAACAAUGUGCUAGUUAUAACCACGACUGUAAGUGGAAAUAAUAUCUACAGUAGCU----------AUUUAGUUACUCUGUAUUGCUUA---- .......(((((((((((.(....(((.........))).).)))))))))))..(((((.......)))))(((((((----------....)))))))............---- ( -26.20, z-score = -2.09, R) >droSim1.chr2L 9870211 102 + 22036055 GAAAGACACAGUCGUGGUCAGAAAAACUAUGUGCUAGUUAUAACCAAGACUGUAAAUGGAAAUAAUAUCUACAGUAGCU----------AUUUAGUUACUCUGUAUUGCUUA---- ..(((..((((((.((((.(....(((((.....))))).).)))).)))))).........((((((....(((((((----------....)))))))..))))))))).---- ( -21.40, z-score = -0.85, R) >consensus GAAAGACACAGUCGUGGUCAGAAAAACAAUGUACUAGCCAUAACCACGACUGUAAAUGGAAAUAAUAUCUACAGUAGAA__________AUUUAGUUACUCUGUAUUGCUUA____ .......(((((((((((........................)))))))))))..................((((((.........................))))))........ (-10.82 = -10.72 + -0.10)

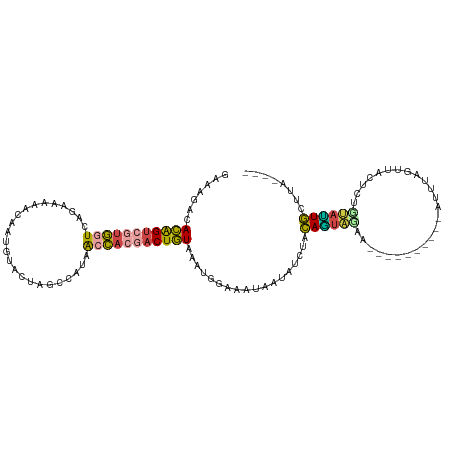
| Location | 10,082,095 – 10,082,206 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 65.10 |
| Shannon entropy | 0.62417 |
| G+C content | 0.34690 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -10.56 |
| Energy contribution | -12.23 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.996127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 10082095 111 - 23011544 ----UAAGCAGUAGAGAGUAACUAAAUAAGUAACUAAAGCUACUGUAGAUAUUAUUUCCACUUACAGUCGUGGCUAUGGCUUGU-CAUUGUUUUCCUGACCACGACUGUGUCUUUC ----...(((((((..(((.(((.....))).)))....)))))))................((((((((((((...(((....-....))).....).)))))))))))...... ( -31.80, z-score = -2.82, R) >droAna3.scaffold_12916 13913125 103 - 16180835 CCUUUAAGAACUUUAAAACAUCCAGCUAACU------UUAUCAUCUAAUUAUCUUAUUAAUUAAAAGUGUUAUUUUUG--UAGUGCAAGAUAC-----ACUUCGUCCUUGUUUCCU ....((((.((....................------........((((((......)))))).((((((...(((((--.....))))).))-----)))).)).))))...... ( -12.10, z-score = -0.93, R) >droEre2.scaffold_4929 10697908 97 - 26641161 ----UAAUCAAUGCA--CUGAGUA-------------UUACAUUGCGGAUACUAUUUCCAUUCACAGUGGUGCUGAUAGCUAGUAUAUCGUUUUUCUGACCACGACUGUGUCUUUC ----........(((--.((....-------------...)).)))(((.......)))...((((((.(((..((((.......))))(((.....)))))).))))))...... ( -20.00, z-score = -0.31, R) >droYak2.chr2L 12766175 99 - 22324452 ----UAAGCAAUACAGUGUGAAUA-------------UUAUAUUGUAGCUUUUAUUUCAAUUUACAGUCGUGGUUAUGGCUAGUAUAUAGUAUUCCUGACCACGACUGUGGCUUUC ----.((((..((((((((((...-------------))))))))))...............((((((((((((((..((((.....)))).....)))))))))))))))))).. ( -34.40, z-score = -5.17, R) >droSec1.super_3 5520304 102 - 7220098 ----UAAGCAAUACAGAGUAACUAAAU----------AGCUACUGUAGAUAUUAUUUCCACUUACAGUCGUGGUUAUAACUAGCACAUUGUUUUUCUGACCACGACUGUGUCUUUC ----(((....((((((((........----------.))).)))))....)))........((((((((((((((.((..(((.....))).)).))))))))))))))...... ( -28.30, z-score = -3.50, R) >droSim1.chr2L 9870211 102 - 22036055 ----UAAGCAAUACAGAGUAACUAAAU----------AGCUACUGUAGAUAUUAUUUCCAUUUACAGUCUUGGUUAUAACUAGCACAUAGUUUUUCUGACCACGACUGUGUCUUUC ----(((....((((((((........----------.))).)))))....)))........(((((((.((((((.(((((.....)))))....)))))).)))))))...... ( -24.90, z-score = -2.70, R) >consensus ____UAAGCAAUACAGAGUAACUAAAU__________AGAUACUGUAGAUAUUAUUUCCAUUUACAGUCGUGGUUAUAGCUAGUACAUAGUUUUUCUGACCACGACUGUGUCUUUC ..............................................................((((((((((((((....................))))))))))))))...... (-10.56 = -12.23 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:30 2011