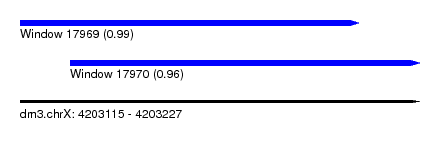
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,203,115 – 4,203,227 |
| Length | 112 |
| Max. P | 0.986692 |

| Location | 4,203,115 – 4,203,210 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.67 |
| Shannon entropy | 0.44977 |
| G+C content | 0.44947 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.30 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.986692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
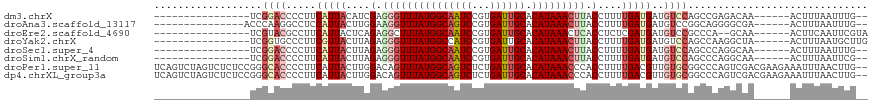
>dm3.chrX 4203115 95 + 22422827 ----------------UCGGACCCCUUCAUUACAUCGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGUCCAGCCGAGACAA------ACUUUAAUUUG-- ----------------..((((....((((((.(..(..((((((((((((((...)))))).))))))))..)..).)))))).))))...........------...........-- ( -25.70, z-score = -3.02, R) >droAna3.scaffold_13117 414248 96 - 5790199 ---------------ACCCAAGGCCUCCAUUACUUGGAAGGUUUAUGGCAGUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGUCCGGCAGGGGCGA------ACUUUAAUUUG-- ---------------.(((...(((..(((((.(..(((((((((((((((((...)))))).)))))....))))))..))))))...)))..)))...------...........-- ( -31.80, z-score = -2.71, R) >droEre2.scaffold_4690 1574750 95 + 18748788 ----------------UCGUACGCCUUCAUUACUCAGAGGCUUUAUGGCAAUCCGUGAUUGCACAUAAACUCACCUCUCGAUGAUGUCCGCCCA--GCAA------ACUUCAAUUCGUA ----------------..((..((...(((((.(((((((.((((((((((((...)))))).))))))....))))).)))))))...))...--))..------............. ( -23.00, z-score = -2.48, R) >droYak2.chrX 17533698 97 - 21770863 ----------------UCGGUGCCCUUCGUUACUUAGAGGGUUUAUGGCCAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGUCCAGCCAAGGCUA------ACUUUAAUGCUUG ----------------..(((.....((((((...((((((((((((((.(((...))).)).)))))))...)))))))))))......))).((((..------........)))). ( -22.70, z-score = -0.61, R) >droSec1.super_4 3644587 95 - 6179234 ----------------UCGGACCCCUUCAUUACUUAGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGUCCAGCCCAGGCAA------ACUUUAAUUUG-- ----------------..((((....((((((...((((((((((((((((((...)))))).)))))))...))))))))))).)))).((....))..------...........-- ( -29.20, z-score = -3.84, R) >droSim1.chrX_random 1577936 95 + 5698898 ----------------UCGGACCCCUUCAUUACUUAGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGUCCAGCCCAGGCAA------ACUUUAAUUCG-- ----------------..((((....((((((...((((((((((((((((((...)))))).)))))))...))))))))))).)))).((....))..------...........-- ( -29.20, z-score = -3.87, R) >droPer1.super_11 2440194 117 + 2846995 UCAGUCUAGUCUCUCCGGGCACCCCUUCAUUACUUGGACAGUUUAUGGCAGUCUCUGAUUGCACAUAAACCCACCUUUUGACGUUGUGCGGCCCAGUCGACGAAGAAAUUUAACUUG-- ....(((.(((...(.((((......(((..(..(((...(((((((((((((...)))))).))))))))))..)..)))((.....)))))).)..)))..)))...........-- ( -29.80, z-score = -1.25, R) >dp4.chrXL_group3a 454918 117 - 2690836 UCAGUCUAGUCUCUCCGGGCACCCCUUCAUUACUUGGACAGUUUAUGGCAGUCUCUGAUUGCACAUAAACCCACCUUUUGACGUUGUGCGGCCCAGUCGACGAAGAAAUUUAACUUG-- ....(((.(((...(.((((......(((..(..(((...(((((((((((((...)))))).))))))))))..)..)))((.....)))))).)..)))..)))...........-- ( -29.80, z-score = -1.25, R) >consensus ________________UCGGACCCCUUCAUUACUUAGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGUCCAGCCCAGGCAA______ACUUUAAUUUG__ ..................((((.....(((((....(.(((((((((((((((...)))))).))))))))).)....)))))..)))).............................. (-16.14 = -17.30 + 1.16)

| Location | 4,203,129 – 4,203,227 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.61 |
| Shannon entropy | 0.52251 |
| G+C content | 0.40510 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -13.44 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
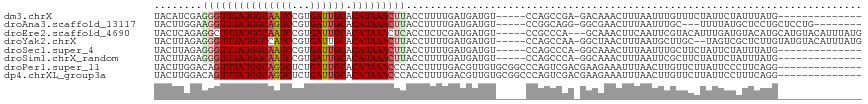
>dm3.chrX 4203129 98 + 22422827 UACAUCGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGU-----CCAGCCGA-GACAAACUUUAAUUUGUUUCUAUUCUAUUUAUG-------------- ..(((((((((((((((((((((...)))))).)))))))).....)))))))....-----......((-((((((......)))))))).............-------------- ( -26.40, z-score = -3.86, R) >droAna3.scaffold_13117 414263 101 - 5790199 UACUUGGAAGGUUUAUGGCAGUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGU-----CCGGCAGG-GGCGAACUUUAAUUUGC---UUUUAUGCUCCUGCUCCUG-------- .....((.(((((((((((((((...)))))).))))))))).))............-----..((((((-((((.............---.....))))))))))....-------- ( -32.27, z-score = -3.04, R) >droEre2.scaffold_4690 1574764 110 + 18748788 UACUCAGAGGCUUUAUGGCAAUCCGUGAUUGCACAUAAACUCACCUCUCGAUGAUGU-----CCGCCCA---GCAAACUUCAAUUCGUACAUUUGAUGUACAUGCAUGUACAUUUAUG ((((((((((.((((((((((((...)))))).))))))....))))).))(((.((-----..((...---))..)).)))....)))(((..((((((((....)))))))).))) ( -27.20, z-score = -2.19, R) >droYak2.chrX 17533712 110 - 21770863 UACUUAGAGGGUUUAUGGCCAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGU-----CCAGCCAA-GGCUAACUUUAAUGCUUGC--UAGUCGCUCUUGUAUGUACAUUUAUG ...((((((((((((((((.(((...))).)).)))))....))))))))).(((((-----.((..(((-(((..(((...........--.))).)).))))..)).))))).... ( -22.90, z-score = -0.01, R) >droSec1.super_4 3644601 98 - 6179234 UACUUAGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGU-----CCAGCCCA-GGCAAACUUUAAUUUGCUUCUAUUCUAUUUAUG-------------- ....(((((((((((((((((((...)))))).)))))))))...............-----.......(-((((((......)))))))....))))......-------------- ( -21.90, z-score = -1.80, R) >droSim1.chrX_random 1577950 98 + 5698898 UACUUAGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGU-----CCAGCCCA-GGCAAACUUUAAUUCGCUUCUAUUCUAUUUAUG-------------- ....(((((((((((((((((((...)))))).))))))).................-----...((...-.)).............))))))...........-------------- ( -21.70, z-score = -1.83, R) >droPer1.super_11 2440224 104 + 2846995 UACUUGGACAGUUUAUGGCAGUCUCUGAUUGCACAUAAACCCACCUUUUGACGUUGUGCGGCCCAGUCGACGAAGAAAUUUAACUUGUUCUUAUUCCCUUCAGG-------------- .....((((((((((((((((((...)))))).))))))).....((((..(((((.((......)))))))..)))).......)))))..............-------------- ( -23.90, z-score = -0.97, R) >dp4.chrXL_group3a 454948 104 - 2690836 UACUUGGACAGUUUAUGGCAGUCUCUGAUUGCACAUAAACCCACCUUUUGACGUUGUGCGGCCCAGUCGACGAAGAAAUUUAACUUGUUCUUAUUCCUUUCAGG-------------- .....((((((((((((((((((...)))))).))))))).....((((..(((((.((......)))))))..)))).......)))))..............-------------- ( -23.90, z-score = -0.99, R) >consensus UACUUAGAGGGUUUAUGGCAAUCCGUGAUUGCACAUAAACUUACCUUUUGAUGAUGU_____CCAGCCGA_GGCAAACUUUAAUUUGUUCCUAUUCUAUUCAUG______________ ........(((((((((((((((...)))))).)))))))))............................................................................ (-13.44 = -13.80 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:08 2011