
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,192,999 – 4,193,068 |
| Length | 69 |
| Max. P | 0.980612 |

| Location | 4,192,999 – 4,193,068 |
|---|---|
| Length | 69 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 60.14 |
| Shannon entropy | 0.77107 |
| G+C content | 0.44735 |
| Mean single sequence MFE | -17.85 |
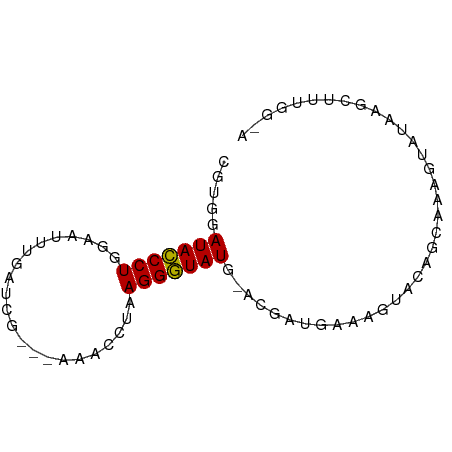
| Consensus MFE | -7.49 |
| Energy contribution | -7.22 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
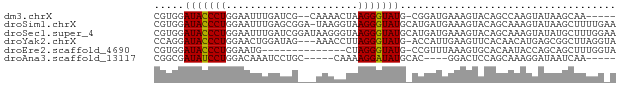
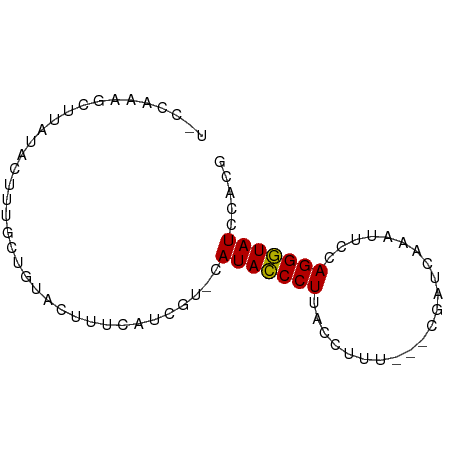
>dm3.chrX 4192999 69 + 22422827 CGUGGAUACCCUGGAAUUUGAUCG--CAAAACUAAGGGUAUG-CGGAUGAAAGUACAGCCAAGUAUAAGCAA----- .(((.(((((((....((((....--))))....))))))).-)((.((......)).))........))..----- ( -15.40, z-score = -1.05, R) >droSim1.chrX 3229485 76 + 17042790 CGUGGAUACCCUGGAAUUUGAGCGGA-UAAGGUAAGGGUAUGCAUGAUGAAAGUACAGCAAAGUAUAAGCUUUUGAA ((((.(((((((....((((......-))))...))))))).))))..((((((((......))....))))))... ( -17.10, z-score = -1.68, R) >droSec1.super_4 3634718 77 - 6179234 CGUGGAUACCCUGGAAUUUGAUCGGAUAAGGGUAAGGGUAUGCAUGAUGAAAGUACAGCAAAGUAUAUGCUUUGGAA ((((.(((((((....((((......))))....))))))).))))............((((((....))))))... ( -20.60, z-score = -2.74, R) >droYak2.chrX 17523395 73 - 21770863 CCAGGAUACCCUGGAACUGGAUAG---AAACCUUAGGGUAUG-ACCAUUGAAGUUCACAACAUGAGCGGCUUAGGUA (((((....)))))..........---..((((.((((....-.))......(((((.....)))))..)).)))). ( -18.40, z-score = -0.72, R) >droEre2.scaffold_4690 1564633 62 + 18748788 CGUGGAUACCCUGGAAUG--------------CUAGGGUAUG-CCGUUUAAAGUGCACAAUACCAGCAGCUUUGGUA ...(((((((((((....--------------))))))))).-))..(((((((((.........))).)))))).. ( -20.40, z-score = -1.57, R) >droAna3.scaffold_13117 394756 63 - 5790199 CGGCGAUAUCCUGGACAAAUCCUGC-----CAAAAGGAUAUGCAC----GGACUCCAGCAAAGGAUAAUCAA----- ....(((((((((..((.(((((..-----....))))).))..)----((...)).....))))).)))..----- ( -15.20, z-score = -1.35, R) >consensus CGUGGAUACCCUGGAAUUUGAUCG___AAACCUAAGGGUAUG_ACGAUGAAAGUACAGCAAAGUAUAAGCUUUGG_A .....(((((((......................))))))).................................... ( -7.49 = -7.22 + -0.28)
| Location | 4,192,999 – 4,193,068 |
|---|---|
| Length | 69 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 60.14 |
| Shannon entropy | 0.77107 |
| G+C content | 0.44735 |
| Mean single sequence MFE | -12.81 |
| Consensus MFE | -5.96 |
| Energy contribution | -5.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
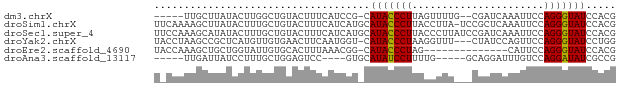
>dm3.chrX 4192999 69 - 22422827 -----UUGCUUAUACUUGGCUGUACUUUCAUCCG-CAUACCCUUAGUUUUG--CGAUCAAAUUCCAGGGUAUCCACG -----............((.((......)).))(-.(((((((....((((--....))))....))))))).)... ( -12.10, z-score = -0.94, R) >droSim1.chrX 3229485 76 - 17042790 UUCAAAAGCUUAUACUUUGCUGUACUUUCAUCAUGCAUACCCUUACCUUA-UCCGCUCAAAUUCCAGGGUAUCCACG ......(((.........)))............((.(((((((.......-..............))))))).)).. ( -9.00, z-score = -1.22, R) >droSec1.super_4 3634718 77 + 6179234 UUCCAAAGCAUAUACUUUGCUGUACUUUCAUCAUGCAUACCCUUACCCUUAUCCGAUCAAAUUCCAGGGUAUCCACG ......((((.......))))............((.(((((((......................))))))).)).. ( -10.65, z-score = -1.93, R) >droYak2.chrX 17523395 73 + 21770863 UACCUAAGCCGCUCAUGUUGUGAACUUCAAUGGU-CAUACCCUAAGGUUU---CUAUCCAGUUCCAGGGUAUCCUGG (((((..((.((....)).))(((((...((((.-...(((....)))..---))))..)))))..)))))...... ( -13.20, z-score = 0.53, R) >droEre2.scaffold_4690 1564633 62 - 18748788 UACCAAAGCUGCUGGUAUUGUGCACUUUAAACGG-CAUACCCUAG--------------CAUUCCAGGGUAUCCACG ....((((.(((.........)))))))....((-.(((((((..--------------......)))))))))... ( -15.80, z-score = -1.13, R) >droAna3.scaffold_13117 394756 63 + 5790199 -----UUGAUUAUCCUUUGCUGGAGUCC----GUGCAUAUCCUUUUG-----GCAGGAUUUGUCCAGGAUAUCGCCG -----..(((.(((((..((..((((((----.(((.((......))-----)))))))))))..)))))))).... ( -16.10, z-score = -0.79, R) >consensus U_CCAAAGCUUAUACUUUGCUGUACUUUCAUCGU_CAUACCCUUACCUUU___CGAUCAAAUUCCAGGGUAUCCACG ....................................(((((((......................)))))))..... ( -5.96 = -5.68 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:06 2011