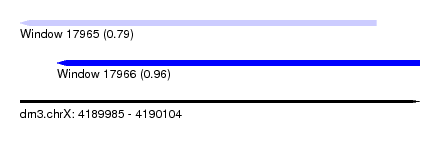
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,189,985 – 4,190,104 |
| Length | 119 |
| Max. P | 0.959271 |

| Location | 4,189,985 – 4,190,091 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.39 |
| Shannon entropy | 0.41230 |
| G+C content | 0.49834 |
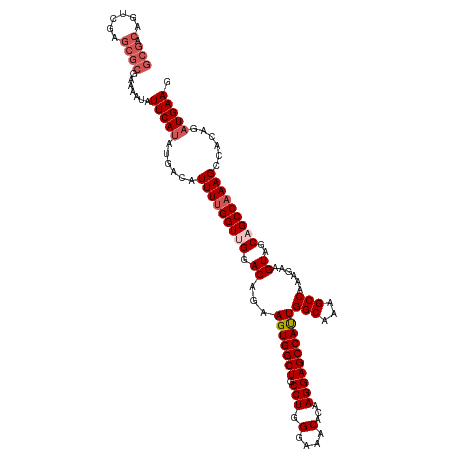
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -18.96 |
| Energy contribution | -20.47 |
| Covariance contribution | 1.51 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4189985 106 - 22422827 CAUAUGACAUUUUGGUUGGA-CAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGA--AGUAGCAGCCAAAGCCACAGAUGAAGCCCA-AGAUGAA (((.((...(((((((((.(-(...(((((((.(((.(....)...))))))))))(((...))).....--.))..)))))))))...)).))).......-....... ( -31.60, z-score = -2.09, R) >droPer1.super_11 2417432 108 - 2846995 GAGAUGAGACUGAAGCUGGAGCUUGAGCUAAGGCC--AAAAGCAGCUGCAGUUGGAAGGCAGACCGAAGACCGCCGCCGGCUGAAAGGCAAAGUGAGUCACAUAAAUUUC ...(((.((((.(.(((.(.((((......)))))--...))).(((.(((((((..(((.(........).))).)))))))...)))....).)))).)))....... ( -32.40, z-score = -0.90, R) >droEre2.scaffold_4690 1561694 106 - 18748788 CAUAUGACAUUUUGGUCGGA-CAGAAGUGGCUGCCUGGGAAACACAAGGAGCCACUGGCAAAGCCAAAGG--AGUAGCAGCCAAAGCCAGAGAUGAAGCCCA-AGAUGAA .......((((((((.(((.-(...(((((((.(((.(....)...))))))))))(((...))).....--.......))).....((....))..).)))-))))).. ( -32.10, z-score = -1.82, R) >droYak2.chrX 17520500 106 + 21770863 CAUAUGACAUUUCGGUCGGA-CAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGA--AGUAGCAGCCAAAGCAAGAGCUGAAGCCCA-AGAUGAA .......(((((.((((...-....(((((((.(((.(....)...))))))))))(((...)))...))--.((..(((((.......).))))..)))).-))))).. ( -29.60, z-score = -1.16, R) >droSec1.super_4 3631706 106 + 6179234 CAUAUGACAUUUUGGUUGGA-CAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGA--AGUAGCAGCCAAAGCCCCAGAUGAAGCCCA-AGAUGAA (((.((...(((((((((.(-(...(((((((.(((.(....)...))))))))))(((...))).....--.))..)))))))))...)).))).......-....... ( -31.60, z-score = -1.81, R) >droSim1.chrX 3226490 106 - 17042790 CAUAUGACAUUUUGGUUGGA-CAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGA--AGUAGCAGCCAAAGCCACAGAUGAAGCCCA-AGAUGAA (((.((...(((((((((.(-(...(((((((.(((.(....)...))))))))))(((...))).....--.))..)))))))))...)).))).......-....... ( -31.60, z-score = -2.09, R) >consensus CAUAUGACAUUUUGGUUGGA_CAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGA__AGUAGCAGCCAAAGCCACAGAUGAAGCCCA_AGAUGAA .........(((((((((.......(((((((.(((.(....)...))))))))))(((...)))............)))))))))........................ (-18.96 = -20.47 + 1.51)

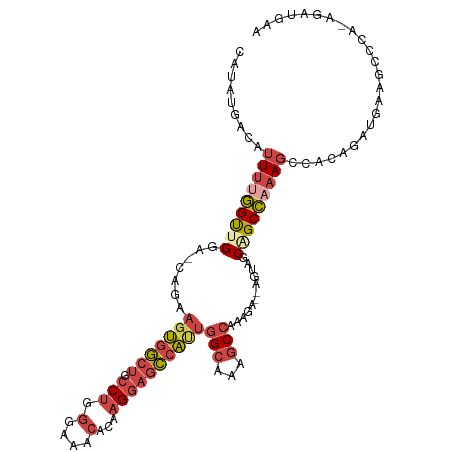
| Location | 4,189,996 – 4,190,104 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.03 |
| Shannon entropy | 0.12491 |
| G+C content | 0.49444 |
| Mean single sequence MFE | -35.56 |
| Consensus MFE | -31.36 |
| Energy contribution | -32.00 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4189996 108 - 22422827 -----------GCGCGAAAAUAUUCAUAUGACAUUUUGGUUGGACAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGAAGUAGCAGCCAAAGCCACAGAUGAAG -----------...........(((((.((...(((((((((.((...(((((((.(((.(....)...))))))))))(((...)))......))..)))))))))...)).))))). ( -34.40, z-score = -3.12, R) >droEre2.scaffold_4690 1561705 119 - 18748788 GCGACAGUCGAGCGCGAAAAUAUUCAUAUGACAUUUUGGUCGGACAGAAGUGGCUGCCUGGGAAACACAAGGAGCCACUGGCAAAGCCAAAGGAGUAGCAGCCAAAGCCAGAGAUGAAG (((.(......)))).......(((((......(((((((.(.((...(((((((.(((.(....)...))))))))))(((...)))......))..).)))))))......))))). ( -35.40, z-score = -1.97, R) >droYak2.chrX 17520511 119 + 21770863 GCGACAGUCGAGCGCGAAAAUAUUCAUAUGACAUUUCGGUCGGACAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGAAGUAGCAGCCAAAGCAAGAGCUGAAG ((.((.(((..((.(((((....((....))..)))))))..)))...(((((((.(((.(....)...))))))))))(((...)))......)).))......(((....))).... ( -31.70, z-score = -0.79, R) >droSec1.super_4 3631717 119 + 6179234 GCGACUGUCGAGCGCGAAAAUAUUCAUAUGACAUUUUGGUUGGACAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGAAGUAGCAGCCAAAGCCCCAGAUGAAG (((.((....))))).......(((((.((...(((((((((.((...(((((((.(((.(....)...))))))))))(((...)))......))..)))))))))...)).))))). ( -38.70, z-score = -2.74, R) >droSim1.chrX 3226501 119 - 17042790 GCGACAGUCGAGCGCGAAAAUAUUCAUAUGACAUUUUGGUUGGACAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGAAGUAGCAGCCAAAGCCACAGAUGAAG (((.(......)))).......(((((.((...(((((((((.((...(((((((.(((.(....)...))))))))))(((...)))......))..)))))))))...)).))))). ( -37.60, z-score = -3.11, R) >consensus GCGACAGUCGAGCGCGAAAAUAUUCAUAUGACAUUUUGGUUGGACAGAAGUGGCUGCCUGGGAAACACAAGGAGCCAUUGGCAAAGCCAAAGAAGUAGCAGCCAAAGCCACAGAUGAAG ......................(((((......(((((((((.((...(((((((.(((.(....)...))))))))))(((...)))......))..)))))))))......))))). (-31.36 = -32.00 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:05 2011