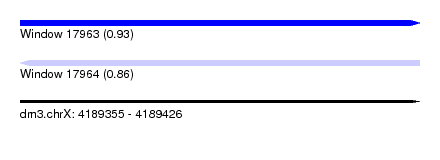
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,189,355 – 4,189,426 |
| Length | 71 |
| Max. P | 0.934800 |

| Location | 4,189,355 – 4,189,426 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 84.95 |
| Shannon entropy | 0.28886 |
| G+C content | 0.43303 |
| Mean single sequence MFE | -19.22 |
| Consensus MFE | -12.55 |
| Energy contribution | -13.03 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
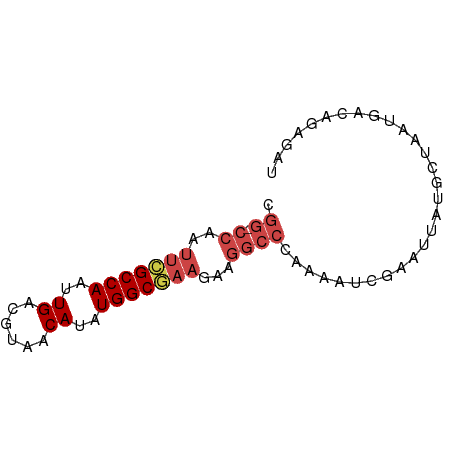
>dm3.chrX 4189355 71 + 22422827 AUCUCUGUCAUUAGCAUAAUUCGUUUUUGGGCCUUCUUCGCCAUAUGUUACGUCAAUUGGCGAAUUGG-CCG ........((..(((.......)))..))((((...(((((((..((......))..)))))))..))-)). ( -20.80, z-score = -3.37, R) >droAna3.scaffold_13117 390536 58 - 5790199 AUCUUUGCCAUUAACAUAAUUGGU-----AAUUUCGUAGGCCAUAUGUUACGGCAAUUGGCCA--------- ....((((((..........))))-----)).......(((((..(((....)))..))))).--------- ( -15.60, z-score = -1.94, R) >droEre2.scaffold_4690 1561030 71 + 18748788 AUCUCUGUCAUUAGCAUAAUUCGAUUUUGGGCCUUCUUUGCCAUAUGUUACGUCAAUUGGCGAAUUGG-CCG .............................((((...(((((((..((......))..)))))))..))-)). ( -17.30, z-score = -1.63, R) >droYak2.chrX 17519846 71 - 21770863 AUCUCUGUCAUUAGCAUAAUUCGAUUUUGGGCCUCCUUCGCCAUAUGUUACGUCAAUUGGCGAAUUGG-CCG .............................((((...(((((((..((......))..)))))))..))-)). ( -19.40, z-score = -2.24, R) >droSec1.super_4 3631088 72 - 6179234 AUCUCUGUCAUUAGCAUAAUUCGAUUUCGGGCCUUCUUCGCCAUAUGUUACGUCAAUUGGCGAAUUGGGCCG .....................((....))(((((..(((((((..((......))..)))))))..))))). ( -22.50, z-score = -3.11, R) >droSim1.chrX 3225875 71 + 17042790 AUCUCUGUCAUUAGCAUAAUUCGAUUUCGGGCCUUCUUCGCCAUAUGUUACGUCAAUUGGCGAAUUGG-CCG .....................((....))((((...(((((((..((......))..)))))))..))-)). ( -19.70, z-score = -2.48, R) >consensus AUCUCUGUCAUUAGCAUAAUUCGAUUUUGGGCCUUCUUCGCCAUAUGUUACGUCAAUUGGCGAAUUGG_CCG ............................((.((...(((((((..((......))..)))))))..)).)). (-12.55 = -13.03 + 0.49)
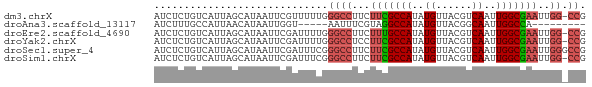
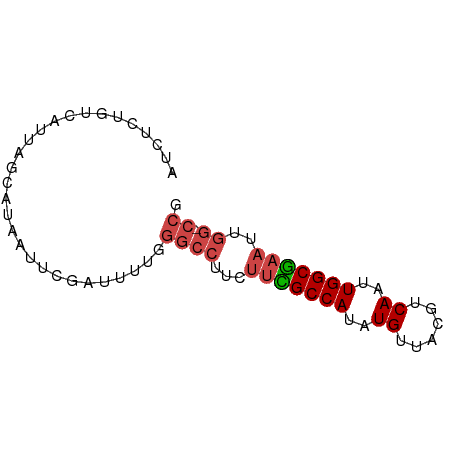
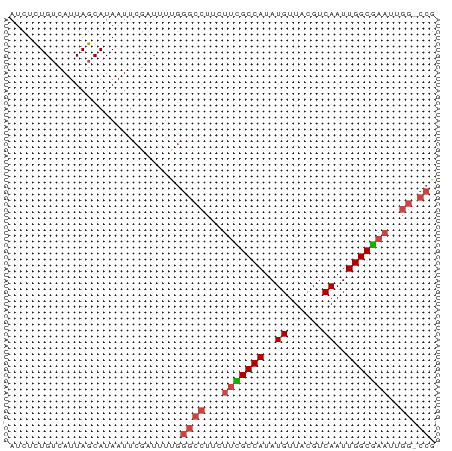
| Location | 4,189,355 – 4,189,426 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 72 |
| Reading direction | reverse |
| Mean pairwise identity | 84.95 |
| Shannon entropy | 0.28886 |
| G+C content | 0.43303 |
| Mean single sequence MFE | -17.30 |
| Consensus MFE | -11.31 |
| Energy contribution | -12.13 |
| Covariance contribution | 0.82 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.864654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4189355 71 - 22422827 CGG-CCAAUUCGCCAAUUGACGUAACAUAUGGCGAAGAAGGCCCAAAAACGAAUUAUGCUAAUGACAGAGAU .((-((..(((((((..((......))..)))))))...))))............................. ( -18.80, z-score = -3.24, R) >droAna3.scaffold_13117 390536 58 + 5790199 ---------UGGCCAAUUGCCGUAACAUAUGGCCUACGAAAUU-----ACCAAUUAUGUUAAUGGCAAAGAU ---------.......((((((((((((((((...........-----.)))..)))))).))))))).... ( -14.20, z-score = -1.77, R) >droEre2.scaffold_4690 1561030 71 - 18748788 CGG-CCAAUUCGCCAAUUGACGUAACAUAUGGCAAAGAAGGCCCAAAAUCGAAUUAUGCUAAUGACAGAGAU .((-((..((.((((..((......))..)))).))...)))).....((...((((....))))..))... ( -13.20, z-score = -0.80, R) >droYak2.chrX 17519846 71 + 21770863 CGG-CCAAUUCGCCAAUUGACGUAACAUAUGGCGAAGGAGGCCCAAAAUCGAAUUAUGCUAAUGACAGAGAU .((-((..(((((((..((......))..)))))))...)))).....((...((((....))))..))... ( -19.10, z-score = -2.39, R) >droSec1.super_4 3631088 72 + 6179234 CGGCCCAAUUCGCCAAUUGACGUAACAUAUGGCGAAGAAGGCCCGAAAUCGAAUUAUGCUAAUGACAGAGAU .((((...(((((((..((......))..)))))))...))))((....))..................... ( -19.40, z-score = -2.60, R) >droSim1.chrX 3225875 71 - 17042790 CGG-CCAAUUCGCCAAUUGACGUAACAUAUGGCGAAGAAGGCCCGAAAUCGAAUUAUGCUAAUGACAGAGAU .((-((..(((((((..((......))..)))))))...))))((....))..................... ( -19.10, z-score = -2.60, R) >consensus CGG_CCAAUUCGCCAAUUGACGUAACAUAUGGCGAAGAAGGCCCAAAAUCGAAUUAUGCUAAUGACAGAGAU .((.((..(((((((..((......))..)))))))...)).))............................ (-11.31 = -12.13 + 0.82)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:03 2011