| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,184,771 – 4,184,879 |
| Length | 108 |
| Max. P | 0.962893 |

| Location | 4,184,771 – 4,184,879 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.39 |
| Shannon entropy | 0.43818 |
| G+C content | 0.35816 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -18.36 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
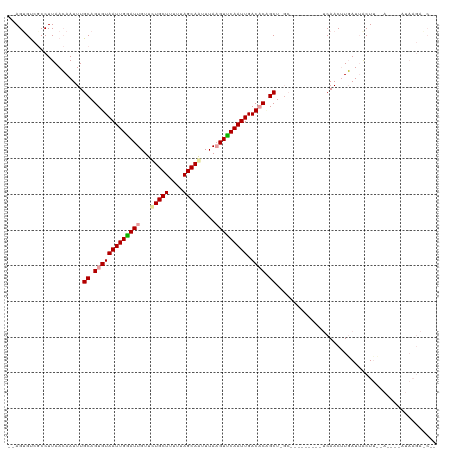
>dm3.chrX 4184771 108 + 22422827 --AGUGUCGCCUUAACAAUUUGCAGUGUAAUUGGAUUGUUCUGCUUUUAAGCAGUUUAAGUUCAAUUGACACUGCUUGC---------UCAACUUGAUUGCUUCGA-GCCAUAAGAACUC --........((((.......((((((((((((((((...(((((....)))))....))))))))).)))))))....---------....(((((.....))))-)...))))..... ( -30.70, z-score = -2.38, R) >droSec1.super_4 3626296 88 - 6179234 --AGGGUAGCCUUAACAAUUUGCAGUGUAAUUGAAUUGUUCUGCUUUUUAGCAAUUUAUGUUCAAUUGACACCGCU-------------CAACAUGAUAACUC----------------- --(((....))).........((.((((((((((((.....((((....))))......)))))))).)))).)).-------------..............----------------- ( -17.70, z-score = -0.89, R) >droYak2.chrX 17513432 120 - 21770863 AAAGUGCCACCUUAAAAAUUUGCAGUGUAAUUGGAUUGUUUUGCUUUUUAGCAAUUUAAGUCCAAUUGACCCAGCUCGCCAACGAUUUUCAACUUUAUUACUUAUAUAGUAUAAGAGCGA ....(((.......((((((.(((((.((((((((((...(((((....)))))....)))))))))).....))).))....))))))...(((...((((.....)))).))).))). ( -23.60, z-score = -1.48, R) >consensus __AGUGUCGCCUUAACAAUUUGCAGUGUAAUUGGAUUGUUCUGCUUUUUAGCAAUUUAAGUUCAAUUGACACAGCU_GC_________UCAACUUGAUUACUU__A____AUAAGA_C__ .....................((.(((((((((((((...(((((....)))))....))))))))).)))).))............................................. (-18.36 = -18.70 + 0.34)


| Location | 4,184,771 – 4,184,879 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.39 |
| Shannon entropy | 0.43818 |
| G+C content | 0.35816 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.80 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
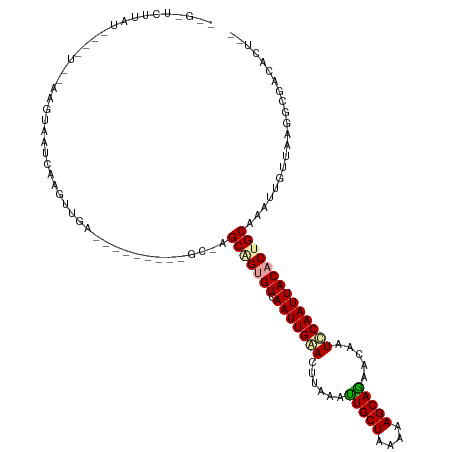
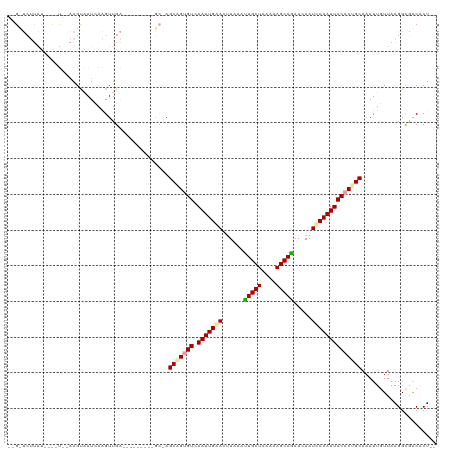
>dm3.chrX 4184771 108 - 22422827 GAGUUCUUAUGGC-UCGAAGCAAUCAAGUUGA---------GCAAGCAGUGUCAAUUGAACUUAAACUGCUUAAAAGCAGAACAAUCCAAUUACACUGCAAAUUGUUAAGGCGACACU-- ..((.((((..((-((((..........))))---------))..(((((((.(((((........(((((....))))).......)))))))))))).......))))))......-- ( -27.66, z-score = -1.82, R) >droSec1.super_4 3626296 88 + 6179234 -----------------GAGUUAUCAUGUUG-------------AGCGGUGUCAAUUGAACAUAAAUUGCUAAAAAGCAGAACAAUUCAAUUACACUGCAAAUUGUUAAGGCUACCCU-- -----------------.((((.((.....)-------------)(((((((.(((((((......(((((....))))).....))))))))))))))..........)))).....-- ( -20.50, z-score = -2.06, R) >droYak2.chrX 17513432 120 + 21770863 UCGCUCUUAUACUAUAUAAGUAAUAAAGUUGAAAAUCGUUGGCGAGCUGGGUCAAUUGGACUUAAAUUGCUAAAAAGCAAAACAAUCCAAUUACACUGCAAAUUUUUAAGGUGGCACUUU ((((.((((((((.....))))..((((((.....(((....)))((...((.(((((((......(((((....))))).....)))))))))...)).))))))))))))))...... ( -22.80, z-score = -0.76, R) >consensus __G_UCUUAU____U__AAGUAAUCAAGUUGA_________GC_AGCAGUGUCAAUUGAACUUAAAUUGCUAAAAAGCAGAACAAUCCAAUUACACUGCAAAUUGUUAAGGCGACACU__ .............................................(((((((.(((((((......(((((....))))).....))))))))))))))..................... (-17.57 = -17.80 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:14:01 2011