
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,164,637 – 4,164,750 |
| Length | 113 |
| Max. P | 0.564250 |

| Location | 4,164,637 – 4,164,750 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.35 |
| Shannon entropy | 0.36485 |
| G+C content | 0.54194 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -20.91 |
| Energy contribution | -22.23 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4164637 113 - 22422827 UUGCGGGGCUCGUCGGCGGCACAGAA---CUCGGGGCUCUGGGUUGU---GCAACGCACAGGACUACAUGCUCACAGCCUCGAGUGCUGUGCAAGCUUAAUUAUAAAAUGUAUGAAGCA- .(((.((.....)).)))((((((.(---(((((((((.((((((((---((...)))))(.....)..))))).)))))))))).))))))..((((...(((.....)))..)))).- ( -45.90, z-score = -3.02, R) >droSim1.chrX 3191887 113 - 17042790 UUGCAGGGCUCGUCGGCAGCACAGGA---CUCGGGGCUCUGGGUUGU---GCAACGCACAGGACUACAUGCUCACAGCCUCGAGUGCUGUGCAAGCUUAAUUAUAAAAUGUAUGAAGCA- .(((..((....)).)))((((((.(---(((((((((.((((((((---((...)))))(.....)..))))).)))))))))).))))))..((((...(((.....)))..)))).- ( -47.90, z-score = -3.53, R) >droSec1.super_4 3611946 113 + 6179234 UUGCAGGGCUCGUCGGCAGCACAGGA---CUCGGGGCUCUGGGUUGU---GCAACGCACAGGACUACAUGCUCACAGCCUCGAGUGCUGUGCAAGCUUAAUUAUAAAAUGUAUGAAGCA- .(((..((....)).)))((((((.(---(((((((((.((((((((---((...)))))(.....)..))))).)))))))))).))))))..((((...(((.....)))..)))).- ( -47.90, z-score = -3.53, R) >droYak2.chrX 17498077 113 + 21770863 UUGCAGGGCUGGUCGGCGGCACAGGA---CUCGGGGCUCUGGGUUGU---GCAACUCACAGGACUACAUGCUCACAGCCGCGAGUGCUGUGCAAGCUUAAUUAUAAAAUGUAUGAAGCC- ......((((....(((.((((((.(---((((.((((.((((((((---(...(.....)...)))).))))).)))).))))).))))))..)))...(((((.....)))))))))- ( -44.00, z-score = -2.09, R) >droEre2.scaffold_4690 1539885 109 - 18748788 UUGCAGGGCUCGUCGGCGGCACAGCA---C----AACCCAGAGUUGU---GCAACUCACAGGACUACAUGCUCACAGCUGCGAGUGCUGUGCAAGCUUAAUUAUAAAAUAUAUGAAGCC- ..((((.(((((.((((((((..(((---(----(((.....)))))---))........(.....).))))....))))))))).))))....((((...((((...))))..)))).- ( -35.10, z-score = -1.09, R) >droAna3.scaffold_13117 5430645 119 - 5790199 UUGCUGGGCUCGUCGGCGGCACAGGAACUCACAGGACUCGCAGCUAUUUGGCAGCUCGUGGCACU-CAUGGCCGUGGACGGGACACGGUUGCAGUUCGAGUGAGUAGAUGCAUGGUGCAU .((((((.....))))))((((..(.((((((.(((((.((((((((..(....)..)))))...-....((((((.......))))))))))))))..)))))).....)...)))).. ( -40.90, z-score = 0.66, R) >consensus UUGCAGGGCUCGUCGGCGGCACAGGA___CUCGGGGCUCUGGGUUGU___GCAACGCACAGGACUACAUGCUCACAGCCGCGAGUGCUGUGCAAGCUUAAUUAUAAAAUGUAUGAAGCA_ (((((.(((..((.....))...............((((.(((((((...(((...............)))..))))))).))))))).)))))((((...(((.....)))..)))).. (-20.91 = -22.23 + 1.31)

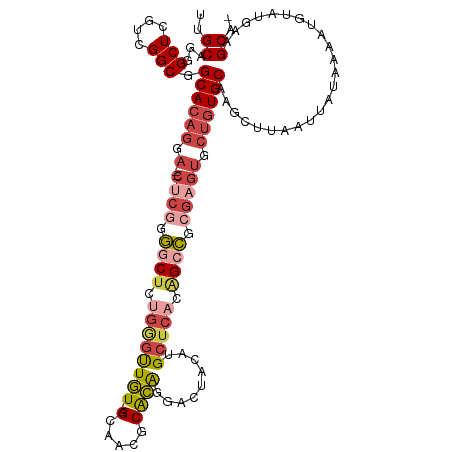
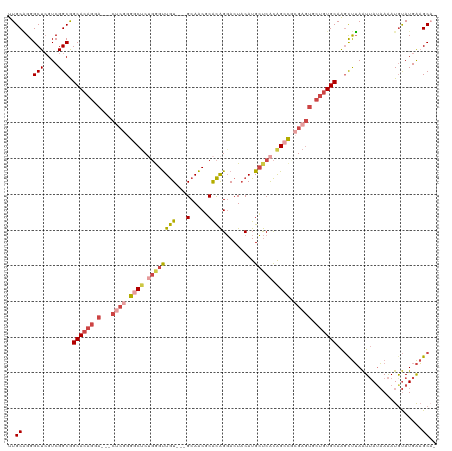
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:59 2011