
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,153,535 – 4,153,631 |
| Length | 96 |
| Max. P | 0.974321 |

| Location | 4,153,535 – 4,153,631 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Shannon entropy | 0.55221 |
| G+C content | 0.49000 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -14.51 |
| Energy contribution | -15.40 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4153535 96 + 22422827 GUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCGCCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAUCGCAGCU-CUGCAAAAUCUAC--- (((((((((((..........)))))))))))..(((.....)))((((((..((((...((((((......)))).))))))-.))))))......--- ( -33.00, z-score = -2.44, R) >droSec1.super_4 3600127 96 - 6179234 GUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCGCCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAUCGCAGGU-CUGCAAAAUCUAC--- (((((((((((..........)))))))))))((((((((..(((((.....)))))..)))((((......))))....)))-..)).........--- ( -32.00, z-score = -2.09, R) >droYak2.chrX 17486738 78 - 21770863 GUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCACCGCACAGGCUUUGUAAAA------------------GCAUCGUUGGU-CUGCAAAAUCUAC--- (((((((((((..........)))))))))))((((((.((..((((.....))------------------))...))))))-..)).........--- ( -24.30, z-score = -2.63, R) >droEre2.scaffold_4690 1528823 96 + 18748788 GUUGUCCUUGCAGACAAUUAAGCAGGGACAAUGCGCCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAGCGCUGGU-CUGCAAAAUCUAC--- (((((((((((..........))))))))))).....(((..(((((.....)))))..))).((((.((..((...))..))-.))))........--- ( -34.90, z-score = -2.02, R) >droAna3.scaffold_13117 347809 97 - 5790199 GUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCGCUGCACUGGCUUUGUAAAAGCUCAUGCAUGCAGACUGCCAGUCCUUCUAUGGCAAUGGUUGC--- (((((((((((..........)))))))))))((((((((..(((((.....)))))..)))).))....(((((.........)))))......))--- ( -34.80, z-score = -1.84, R) >dp4.chrXL_group3a 381691 97 - 2690836 ---GUCCUUGCAGACAAUUAAGCAAGGACAAUGCCAGGCAGCAGCUUUAUAAAUGCUCAUGAAUGCACACAGAAGCCGAAGGACUCGACCCCGCCCCUGG ---((((((((..........))))))))....(((((..((.(((((.....(((........)))....)))))(((.....))).....)).))))) ( -30.10, z-score = -2.61, R) >droPer1.super_11 2371238 97 + 2846995 ---GUCCUUGCAGACAAUUAAGCAAGGACAAUGCCAGGCAGCAGCUUUAUAAAUGCUCAUGAAUGCACACAGAAGCCGAAGGACUCGACCCCGCCCCUGG ---((((((((..........))))))))....(((((..((.(((((.....(((........)))....)))))(((.....))).....)).))))) ( -30.10, z-score = -2.61, R) >droGri2.scaffold_14853 6450164 92 - 10151454 GUUGUCCUUGCAGACAAUUAAGCAGGAACAAUGCA--GCACAAGUCUUAUAAAUGCUCAUGCAUGCC-ACUGCGAGUAA--CUAUUGCAACACCCAC--- (((((.(((((..........))))).)))))(((--(((..(((.........)))..))).))).-..(((((....--...)))))........--- ( -18.90, z-score = 0.20, R) >consensus GUUGUCCUUGCAGACAAUUAAGCAAGGACAAUGCGCCGCACAGGCUUUGUAAAAGCUCAUGCAUGCACACUGGCAGCGCAGGU_CUGCAAAAUCUAC___ (((((((((((..........)))))))))))((...((....)).........((........))....................))............ (-14.51 = -15.40 + 0.89)

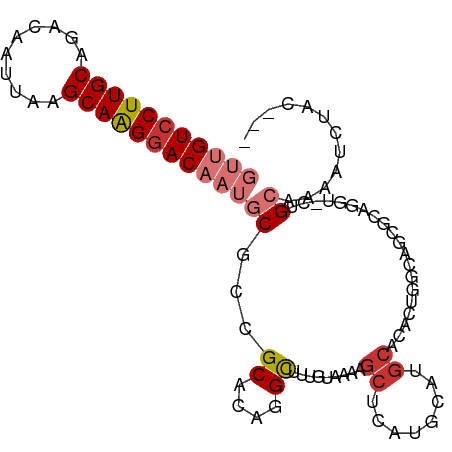
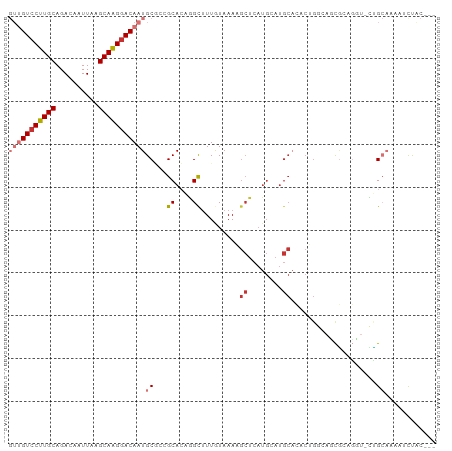
| Location | 4,153,535 – 4,153,631 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 72.50 |
| Shannon entropy | 0.55221 |
| G+C content | 0.49000 |
| Mean single sequence MFE | -33.76 |
| Consensus MFE | -13.03 |
| Energy contribution | -14.36 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4153535 96 - 22422827 ---GUAGAUUUUGCAG-AGCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGGCGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAAC ---.......(((((.-...)))))....(((((((.(((((..((((.....))))..))))))))))))((((((((..........))))))))... ( -36.90, z-score = -2.44, R) >droSec1.super_4 3600127 96 + 6179234 ---GUAGAUUUUGCAG-ACCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGGCGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAAC ---...(.((((((((-((..((((.(.((((((((.(((((..((((.....))))..))))))))))))).).))))......)))))))))).)... ( -38.90, z-score = -3.63, R) >droYak2.chrX 17486738 78 + 21770863 ---GUAGAUUUUGCAG-ACCAACGAUGC------------------UUUUACAAAGCCUGUGCGGUGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAAC ---........(((..-(((.(((..((------------------((.....)))).)))..))))))((((((((((..........)))))))))). ( -23.50, z-score = -2.05, R) >droEre2.scaffold_4690 1528823 96 - 18748788 ---GUAGAUUUUGCAG-ACCAGCGCUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGGCGCAUUGUCCCUGCUUAAUUGUCUGCAAGGACAAC ---...(.((((((((-((.((((..(.((((((((.(((((..((((.....))))..))))))))))))).)..)))).....)))))))))).)... ( -37.90, z-score = -2.80, R) >droAna3.scaffold_13117 347809 97 + 5790199 ---GCAACCAUUGCCAUAGAAGGACUGGCAGUCUGCAUGCAUGAGCUUUUACAAAGCCAGUGCAGCGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAAC ---((....(((((((.........)))))))..((.(((((..((((.....))))..))))))))).((((((((((..........)))))))))). ( -37.00, z-score = -2.89, R) >dp4.chrXL_group3a 381691 97 + 2690836 CCAGGGGCGGGGUCGAGUCCUUCGGCUUCUGUGUGCAUUCAUGAGCAUUUAUAAAGCUGCUGCCUGGCAUUGUCCUUGCUUAAUUGUCUGCAAGGAC--- (((((((((((((((((...)))))))))...((((........))))..........))).)))))....((((((((..........))))))))--- ( -34.80, z-score = -1.48, R) >droPer1.super_11 2371238 97 - 2846995 CCAGGGGCGGGGUCGAGUCCUUCGGCUUCUGUGUGCAUUCAUGAGCAUUUAUAAAGCUGCUGCCUGGCAUUGUCCUUGCUUAAUUGUCUGCAAGGAC--- (((((((((((((((((...)))))))))...((((........))))..........))).)))))....((((((((..........))))))))--- ( -34.80, z-score = -1.48, R) >droGri2.scaffold_14853 6450164 92 + 10151454 ---GUGGGUGUUGCAAUAG--UUACUCGCAGU-GGCAUGCAUGAGCAUUUAUAAGACUUGUGC--UGCAUUGUUCCUGCUUAAUUGUCUGCAAGGACAAC ---(((((((.........--.)))))))...-.(((.(((..(((........).))..)))--))).((((((.(((..........))).)))))). ( -26.30, z-score = -0.20, R) >consensus ___GUAGAUUUUGCAA_ACCUGCGAUGCCAGUGUGCAUGCAUGAGCUUUUACAAAGCCUGUGCGGCGCAUUGUCCUUGCUUAAUUGUCUGCAAGGACAAC ...........(((....................((........)).........((....))...)))((((((((((..........)))))))))). (-13.03 = -14.36 + 1.33)
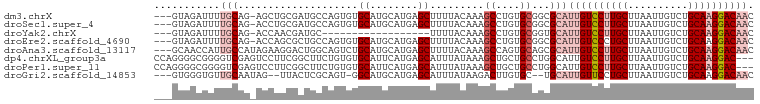
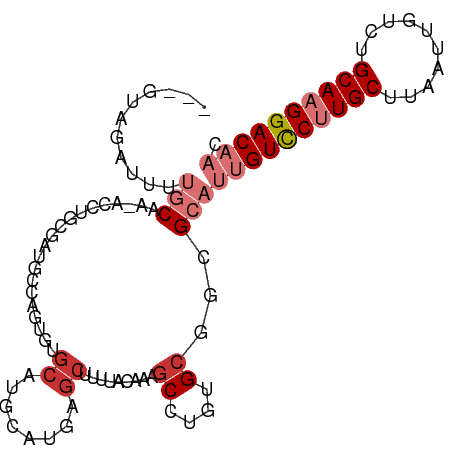
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:55 2011