| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,152,332 – 4,152,402 |
| Length | 70 |
| Max. P | 0.985444 |

| Location | 4,152,332 – 4,152,402 |
|---|---|
| Length | 70 |
| Sequences | 12 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 80.98 |
| Shannon entropy | 0.41306 |
| G+C content | 0.41154 |
| Mean single sequence MFE | -13.29 |
| Consensus MFE | -8.82 |
| Energy contribution | -8.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985444 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chrX 4152332 70 - 22422827 GCCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAGCACAUUUAUUGCUACAACCA--AUCCCAC-- ((..((..((((((((...))))))))..))(-(((....)).)).........))........--.......-- ( -12.50, z-score = -2.17, R) >droSim1.chrX 3118309 70 - 17042790 GCCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAGCACAUUUAUUGCUACAACCA--ACCCCAC-- ((..((..((((((((...))))))))..))(-(((....)).)).........))........--.......-- ( -12.50, z-score = -2.29, R) >droSec1.super_4 3598756 70 + 6179234 GGCAGCUCUUAAAUAUUUCAUAUUUAAUUGCGCCGCUUACGC-GCACAUUUAUUGCUACAACCA--ACCCCAC-- (((.((..((((((((...))))))))..)))))((....))-(((.......)))........--.......-- ( -13.30, z-score = -2.51, R) >droYak2.chrX 17485513 70 + 21770863 GCCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAGCACAUUUAUUCCUACAACCA--ACCCCUC-- ....((..((((((((...))))))))..))(-(((....)).))...................--.......-- ( -11.70, z-score = -3.33, R) >droEre2.scaffold_4690 1527602 70 - 18748788 GCCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAGCACAUUUAUUGCUACAACCA--ACCCCUC-- ((..((..((((((((...))))))))..))(-(((....)).)).........))........--.......-- ( -12.50, z-score = -2.31, R) >droAna3.scaffold_13117 346478 67 + 5790199 ---AGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAGCACAUUUAUUGCUAUAACCA--CCCCCUC-- ---.((..((((((((...))))))))..)).-.((....))((((.......)))).......--.......-- ( -12.10, z-score = -2.58, R) >dp4.chrXL_group3a 379532 72 + 2690836 GCUAGCUCUUAAAUAUUUCAUAUUUAAUUGCA-CGCGUAAGCAGCACAUUUAUUGCUACAACCAGGAACCCAC-- (((.((..((((((((...))))))))..)).-.((....)))))............................-- ( -12.80, z-score = -1.82, R) >droPer1.super_11 2369085 72 - 2846995 GCUAGCUCUUAAAUAUUUCAUAUUUAAUUGCA-CGCGUAAGCAGCACAUUUAUUGCUACAACCAGGAACCCAC-- (((.((..((((((((...))))))))..)).-.((....)))))............................-- ( -12.80, z-score = -1.82, R) >droWil1.scaffold_181150 2728278 70 + 4952429 GGCAGCUCUUAAAUAUUUCAUAUUUAAUUUCA-CGCGUAAGCAGCACAUUUAUUGCCACGUUUU--ACCCCUC-- (((((...((((((((...)))))))).....-.((....))..........))))).......--.......-- ( -12.30, z-score = -2.93, R) >droVir3.scaffold_12928 6857343 72 - 7717345 GGCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAUUGCAACCCU--CUAACACUGCGUACCCCUGA .((((...((((((((...))))))))(((((-.((....))..)))))....--......)))).......... ( -14.20, z-score = -1.14, R) >droMoj3.scaffold_6328 2526522 64 - 4453435 GGCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAUUGCAACCCU--CUAACAUUGCCA-------- ((((((..((((((((...))))))))..))(-(((....))...))......--.......)))).-------- ( -16.30, z-score = -2.65, R) >droGri2.scaffold_14853 6445048 71 + 10151454 GGCAGCUUUUAAAUAUUUCAUAUUUAAUUGCG-CGCGUAAGCAAUGCAACCCU--CUACGAACGCCUCCCGCCA- (((.((..((((((((...))))))))..))(-(((....))...))......--...............))).- ( -16.50, z-score = -1.98, R) >consensus GCCAGCUCUUAAAUAUUUCAUAUUUAAUUGCG_CGCGUAAGCAGCACAUUUAUUGCUACAACCA__ACCCCAC__ ....((..((((((((...))))))))..))...((....))................................. ( -8.82 = -8.90 + 0.08)

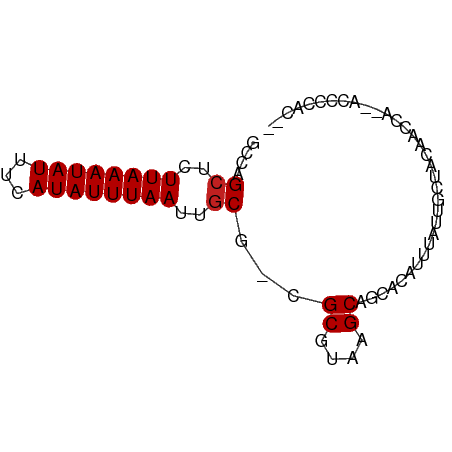
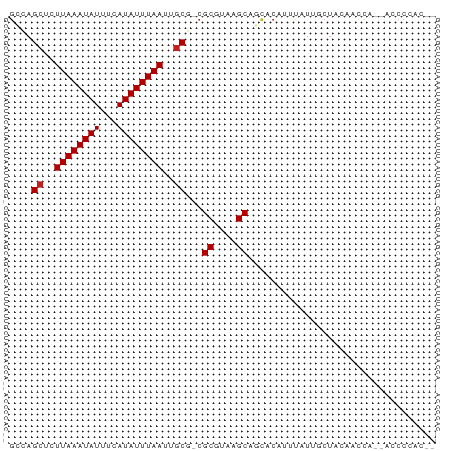
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:54 2011