| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,151,536 – 4,151,647 |
| Length | 111 |
| Max. P | 0.897544 |

| Location | 4,151,536 – 4,151,647 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 64.10 |
| Shannon entropy | 0.67002 |
| G+C content | 0.49106 |
| Mean single sequence MFE | -18.56 |
| Consensus MFE | -7.75 |
| Energy contribution | -7.69 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.897544 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
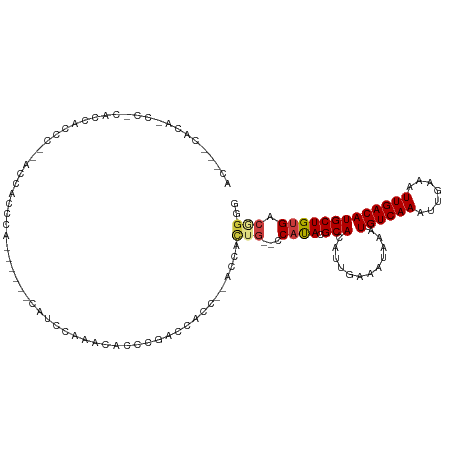
>dm3.chrX 4151536 111 + 22422827 ACCCUCACCACCCUACCACCCACCACAUCCACA---UACACCCAUUGGACCACCAUC----ACUGC---CAUACGCACAUUGAAAUAAAUGUCAAAUUGAAAUUGACAUGCUGUGACGGGG .(((((((...................((((..---.........))))......((----(.(((---.....)))...))).....(((((((.......)))))))...)))).))). ( -18.40, z-score = -0.64, R) >droSim1.chrX 3117539 102 + 17042790 --------ACCCUCACCACCCU-ACCACCCA------CAUCCACCCAUUGGACCACC--AUCACUG--CCAUACGCACAUUGAAAUAAAUGUCAAAUUGAAAUUGACAUGCUGUGACGGGG --------.(((((((......-........------..((((.....)))).....--.(((.((--(.....)))...))).....(((((((.......)))))))...)))).))). ( -19.40, z-score = -1.08, R) >droSec1.super_4 3597987 94 - 6179234 -----------------ACCCUCACCACCCA------CAUCCACCCAUUGGACCACC--AUCACUG--CCAUACGCACAUUGAAAUAAAUGUCAAAUUGAAAUUGACAUGCUGUGACGGGG -----------------.(((((((......------..((((.....)))).....--.(((.((--(.....)))...))).....(((((((.......)))))))...)))).))). ( -19.40, z-score = -1.01, R) >droYak2.chrX 17484712 88 - 21770863 -----------------------ACCCCCCA------CAUCCACCCGUUUGAUCACC--ACCACUG--CCAUACGCACAUUGAAAUAAAUGUCAAAUUGAAAUUGACAUGCUGUGGCGAGG -----------------------.....((.------((..(....)..))......--.....((--(((((.(((............((((((.......)))))))))))))))).)) ( -15.50, z-score = -0.48, R) >dp4.chrXL_group3a 378301 120 - 2690836 ACCCACACAGCCGCAGCUCCCCAGCCGCCCAGCCGCCCACUGGAACACCCAACCAUUGAACCACUGAACCACACGCACAAUAAAAUAAAUGUCAAAUUAAAAUUGACAUGCUGUGACAGA- .....((((((.((.(((....)))...((((.......))))...............................))............(((((((.......))))))))))))).....- ( -23.30, z-score = -2.98, R) >droPer1.super_11 2367741 112 + 2846995 ACCCACACAGCCGCAGCUCCCCAGCCGCCCA--------CUGGAACACCCAACCAUUGAACCACUGAACCACACGCACAAUAAAAUAAAUGUCAAAUUAAAAUUGACAUGCUGUGACAGA- .....((((((.((.(((....)))....((--------.(((.........))).))................))............(((((((.......))))))))))))).....- ( -21.10, z-score = -3.00, R) >droWil1.scaffold_181150 2220978 105 + 4952429 ACGUCCACAAUCAUACC------CACACCCACA---CCCAUCCGACACCCUGCCACC----CCCCG---CCUACGCACAGUAAAAUGAAUGUCAAAUUGAAAUUGACAUGCUGUGACAGAG ..(((.(((.((((...------..........---...............((....----....)---).(((.....)))..))))(((((((.......)))))))..)))))).... ( -12.80, z-score = -0.74, R) >consensus AC___CACA_CC_CACCACCC__ACCACCCA______CAUCCAAACACCCGACCACC__ACCACUG__CCAUACGCACAUUGAAAUAAAUGUCAAAUUGAAAUUGACAUGCUGUGACGGGG .....................................................................((((.(((............((((((.......)))))))))))))...... ( -7.75 = -7.69 + -0.06)

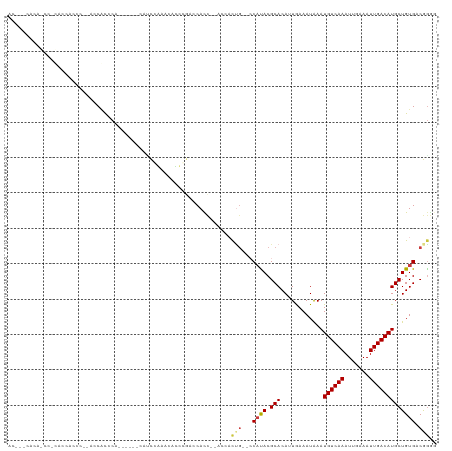
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:53 2011