| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,138,257 – 4,138,316 |
| Length | 59 |
| Max. P | 0.724120 |

| Location | 4,138,257 – 4,138,316 |
|---|---|
| Length | 59 |
| Sequences | 11 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 64.10 |
| Shannon entropy | 0.76107 |
| G+C content | 0.49140 |
| Mean single sequence MFE | -11.22 |
| Consensus MFE | -5.19 |
| Energy contribution | -4.81 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 2.56 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4138257 59 - 22422827 GAACCAUUUCCGGAGUUGCC-CCUCGACUUAGGAAAUAUCGACAACAACAGAAUCCCCAA- .....((((((.((((((..-...)))))).)))))).......................- ( -11.20, z-score = -1.86, R) >droSim1.chrX 3104634 59 - 17042790 GGAUUCUUUCCAGAGUUGCC-UCUCGAUUUAGGAAAUAUCGACAACAAUAUUAUCCCCAA- .(((..(((((.((((((..-...)))))).))))).)))....................- ( -9.30, z-score = -0.69, R) >droYak2.chrX 17471188 59 + 21770863 GAACCAUUUCCGCAGUUACC-CCUUAAUUUGGGAAAUAUCGACAAUAAUAGAAUCCCCAA- .....((((((.(((.....-.......))))))))).......................- ( -5.70, z-score = -0.00, R) >droEre2.scaffold_4690 1513255 59 - 18748788 GAACCAUUUCCACAAUUGCC-CCUCGAUUUAGGAAAUAUCGACAAUAAUAGAAUCCCUAA- .....((((((..(((((..-...)))))..)))))).......................- ( -6.10, z-score = -1.13, R) >droAna3.scaffold_13117 323498 59 + 5790199 GAACCCAUUCCAGGUACCGC-CCUGGAUCUGGGUAAUAUAGAUAACCUAAGGAUUCCCGA- ..(((((.((((((......-))))))..)))))................((....))..- ( -18.10, z-score = -1.79, R) >dp4.chrXL_group3a 363972 59 + 2690836 GAACCACAGCUCGGUCUGCC-CCUCGAUCUGGGUAACAUACACAAUGUGCGCAUUUCCCA- ..(((.(((.((((......-..)))).)))))).((((.....))))............- ( -10.40, z-score = 0.51, R) >droPer1.super_11 2352272 61 - 2846995 GAACCACAGCUCGGUCUGCCUCCUCGAUCUGGGUAACAUACACAAUGUGCGCAUUUCCCGA ..(((.(((.((((.........)))).)))))).((((.....))))............. ( -9.60, z-score = 1.08, R) >droWil1.scaffold_181150 2697784 59 + 4952429 GAACCUCUGCCUGGCUUGCC-CCUGAAUCUAGGCAACAUUCAUAAUCAACGGAUACCACA- (((....(((((((.((...-....)).)))))))...)))...................- ( -9.50, z-score = -0.40, R) >droVir3.scaffold_12928 6838083 59 - 7717345 GAGCCGCUGCCAGAUCUACC-CCUGCGAUUGGGCCACAUCGACAACCAGCGCAUUGAUCA- ..((.((((...(((....(-((.......)))....)))......))))))........- ( -11.90, z-score = 0.33, R) >droMoj3.scaffold_6328 2506328 59 - 4453435 GAGCCCCUGCCAGAGUUGCC-UCUGCGCCUGGGCAACAUCGAGAACCAGCGCAUCGAUCA- ..((((..(((((((....)-)))).))..))))...(((((...........)))))..- ( -20.20, z-score = -1.41, R) >droGri2.scaffold_14853 6426916 59 + 10151454 GAGCCACUGCCAGAGUUGCC-ACUUAAUUUGGGCAACAUCGACAAUCAUCGCAUUAUCCC- .......(((..((((((((-.(.......))))))).))((......))))).......- ( -11.40, z-score = -0.99, R) >consensus GAACCACUGCCAGAGUUGCC_CCUCGAUUUGGGAAACAUCGACAAUCAGCGCAUCCCCCA_ ........(((.((((((......)))))).)))........................... ( -5.19 = -4.81 + -0.38)

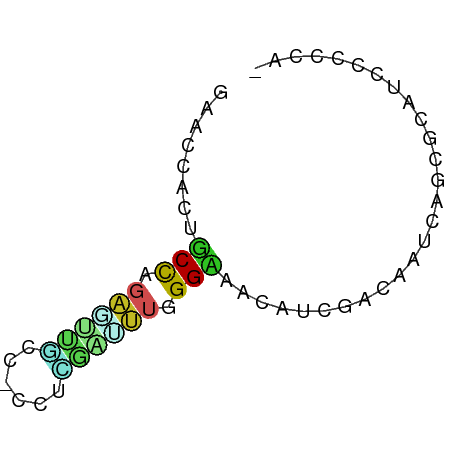

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:51 2011