
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,993,236 – 9,993,348 |
| Length | 112 |
| Max. P | 0.860267 |

| Location | 9,993,236 – 9,993,348 |
|---|---|
| Length | 112 |
| Sequences | 15 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Shannon entropy | 0.58559 |
| G+C content | 0.50641 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.09 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.860267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9993236 112 + 23011544 ----AAUCCUUCCUCCUCCCUCUUCUCCUUGCAGAUGCCGUACCUGAAUCCAGCCCGCUCCCUGGGUCCUUCGUUUGUGCUUAACAAAUGGGACAGCCAUUGGGUGUACUGGUUCG ----..........................((....)).......(((.((((..(((((..(((((((..(((((((.....)))))))))))..)))..)))))..))))))). ( -29.40, z-score = -1.16, R) >droWil1.scaffold_180708 12303987 116 + 12563649 AACUUAUAUUUUUCCCUAUUAAAUUUAAUUACAGAUGCCUUACUUGAAUCCAGCGCGCUCUUUGGGUCCAUCGUUUGUGUUGAACAAAUGGGACAAUCAUUGGGUAUAUUGGUUUG ...............................(((((((((....(((..((((........))))((((..(((((((.....)))))))))))..)))..)))))).)))..... ( -23.00, z-score = -0.24, R) >droSim1.chr2L 9777769 112 + 22036055 ----AAUCAUUUCUCCUCGCUCUUCUCCUUGCAGAUGCCAUACCUGAAUCCAGCCCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAGCCAUUGGGUGUACUGGUUCG ----..............(((((.(.....).))).)).......(((.((((..(((((..(((((((..(((((((.....)))))))))))..)))..)))))..))))))). ( -31.60, z-score = -1.58, R) >droSec1.super_3 5432878 112 + 7220098 ----AAUCAUUUCUCCUCCCUCUUCUCCUUGCAGAUGCCAUACCUGAAUCCAGCCCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAGCCAUUGGGUGUACUGGUUCG ----..........................((....)).......(((.((((..(((((..(((((((..(((((((.....)))))))))))..)))..)))))..))))))). ( -29.40, z-score = -1.10, R) >droYak2.chr2L 12676931 109 + 22324452 -------AAUCCUCCCGCCUUCUUCUCCUUGCAGAUGCCAUACCUGAAUCCGGCCCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAGCCACUGGGUGUACUGGUUCG -------.........((..(((.(.....).))).)).......(((.((((..(((((..(((((((..(((((((.....)))))))))))..)))..)))))..))))))). ( -30.50, z-score = -0.67, R) >droEre2.scaffold_4929 10606426 109 + 26641161 -------AAUCCUCCCUCCUUCUCCUCCUUGCAGAUGCCAUACCUGAAUCCGGCCCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAGCCACUGGGUGUACUGGUUCG -------.......................((....)).......(((.((((..(((((..(((((((..(((((((.....)))))))))))..)))..)))))..))))))). ( -28.70, z-score = -0.41, R) >droAna3.scaffold_12916 13824940 107 + 16180835 ---------AAUGCUUAUCCUUGCCUCCUUUCAGAUGCCCUACUUGAAUCCUGCCCGCUCCCUGGGUCCUUCGUUUGUGCUGAACAAAUGGGACCACCACUGGGUGUACUGGUUCG ---------.............((.((......)).)).......(((((..(..(((((....(((((..(((((((.....))))))))))))......)))))..).))))). ( -25.90, z-score = -0.40, R) >anoGam1.chr3R 17672530 110 + 53272125 ------UCUUUUCCUCAUUCAUUCCCAUUUGCAGAUGCCCUAUCUGAACCCGGCCCGCUCGCUCGGUCCCUCGUUUGUGCUGAACAAAUGGGACAACCACUGGGUGUACUGGGUCG ------.........................((((((...)))))).((((((..((((((....(((((..((((((.....)))))))))))......))))))..)))))).. ( -33.00, z-score = -1.57, R) >triCas2.chrUn_11 34894 107 + 1365317 ---------UUCAUUAAUCAUUUUUUGAAUUUAGAUGCCGCACUUGAACCCAGCGCGUUCGCUGGGGCCGUCCUUUGUUUUGAAAAAAUGGGACAACCACUGGGUUUAUUGGCUGG ---------((((..((....))..))))........((.........(((((((....)))))))((((((((....(((....))).)))))((((....))))....))).)) ( -29.90, z-score = -0.97, R) >droGri2.scaffold_15252 3701766 96 + 17193109 ------------------UUGA--UCCAUUGCAGAUGCCCUAUCUGAAUCCAGCUCGCUCUUUGGGUCCAUCGUUUGUGCUUAACAAAUGGGAUAAUCAUUGGGUAUAUUGGUUCG ------------------..((--.(((......((((((....(((((((.(((((.....)))))....(((((((.....)))))))))))..)))..))))))..))).)). ( -25.50, z-score = -1.00, R) >droVir3.scaffold_12723 1846554 88 - 5802038 ----------------------------UUGCAGAUGCCAUAUCUGAACCCGGCGCGUUCUUUGGGUCCAUCGUUUGUGCUCAACAAAUGGGACAACCAUUGGGUAUACUGGUUCG ----------------------------...((((((...))))))...((((.(..(((..(((((((..(((((((.....)))))))))))..)))..)))..).)))).... ( -24.60, z-score = -0.38, R) >apiMel3.GroupUn 17128220 98 + 399230636 ------------------CUUUAUCUAAUUUCAGAGCCCGUUUUUAAAUCCCGCCCGAGCACUCGGUCCAGCGUUCGUCAGCAACCGUUGGGACAACCAUUGGGUUUACUGGGUGG ------------------.................(((((....(((((((...((((....))))(((((((...(....)...))))))).........))))))).))))).. ( -23.90, z-score = 0.32, R) >droMoj3.scaffold_6500 14778446 105 + 32352404 ---------UUUUCCCUCUUC--UUCCAUUGCAGAUGCCCUAUCUGAAUCCGGCGCGCUCCUUGGGUCCAUCGUUUGUGCUGAACAAAUGGGAUAAUCAUUGGGUGUAUUGGUUUG ---------............--........((((((...))))))...((((..(((((..(((((((..(((((((.....)))))))))))..)))..)))))..)))).... ( -26.30, z-score = -0.17, R) >dp4.chr4_group3 11505811 107 - 11692001 ---------UUCUCAUUCGAUUCUUUCCUUGCAGAUGCCCUACCUGAAUCCGGCUCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAACCAUUGGGUCUACUGGUUCG ---------.(((((...((.....))..)).)))..........(((.((((...((((..(((((((..(((((((.....)))))))))))..)))..))))...))))))). ( -25.30, z-score = -0.22, R) >droPer1.super_1 8621834 107 - 10282868 ---------UUCUCAUUCGAUUCUCUCCUUGCAGAUGCCCUACCUGAAUCCGGCUCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAACCAUUGGGUCUACUGGUUCG ---------.....................((....)).......(((.((((...((((..(((((((..(((((((.....)))))))))))..)))..))))...))))))). ( -24.60, z-score = 0.04, R) >consensus _________UUCUCCCUCCUUUUUCUCCUUGCAGAUGCCCUACCUGAAUCCGGCCCGCUCCCUGGGUCCUUCGUUUGUGCUCAACAAAUGGGACAACCAUUGGGUGUACUGGUUCG ...............................((((((((..........((((........))))((((..(((((((.....)))))))))))........))))).)))..... (-19.24 = -19.09 + -0.15)
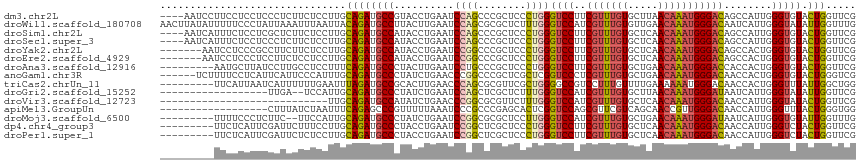

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:26 2011