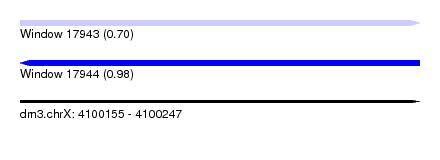
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,100,155 – 4,100,247 |
| Length | 92 |
| Max. P | 0.980770 |

| Location | 4,100,155 – 4,100,247 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 65.70 |
| Shannon entropy | 0.63035 |
| G+C content | 0.55347 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -11.21 |
| Energy contribution | -11.09 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.702335 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
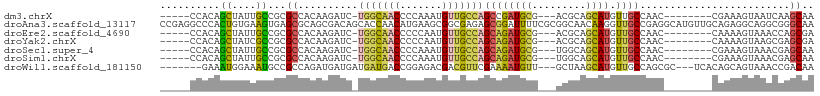
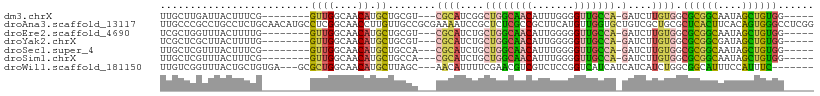
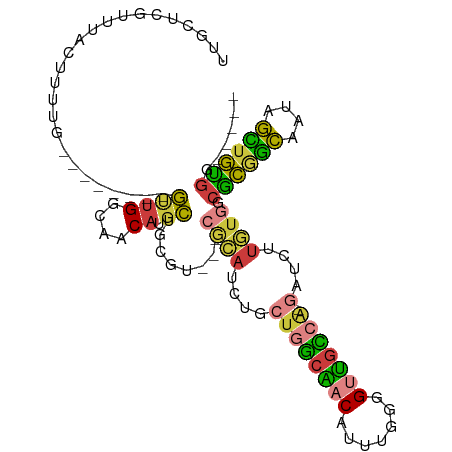
>dm3.chrX 4100155 92 + 22422827 -----CCACAGCUAUUGCCGCGCCACAAGAUC-UGGCAACCCCAAAUGUUGCCAGCCGAUGCG---ACGCAGCAUGUUGCCAAC--------CGAAAGUAAUCAAGCAA -----...((((...(((.(((.(.((....(-(((((((.......))))))))....)).)---.))).))).)))).....--------(....)........... ( -25.60, z-score = -1.69, R) >droAna3.scaffold_13117 273503 109 - 5790199 CCGAGGCCCACUGUGAAGUGAGCGCAGCGACAGCACCAACAUGAAGCGGCGAGAGCGGAUUUUCGCGGCAACAAGGUUGCCGAGGCAUGUUGCAGAGGCAGGCGGGCAA .....((((.((((...((....)).((((((((..(...........(((((((....)))))))((((((...)))))))..)).))))))....))))..)))).. ( -41.30, z-score = -0.96, R) >droEre2.scaffold_4690 1470417 92 + 18748788 -----CCACAGCUAUUGCCGCGCCACAAGAUC-UGGCAACCCCCAAUGUUGCCAGCAGAUGCG---ACGCAGCAUGUUGCCAAC--------CAAAAGUAAACCAGCGA -----...((((...(((.(((.(.((....(-(((((((.......))))))))....)).)---.))).))).)))).....--------................. ( -25.10, z-score = -1.66, R) >droYak2.chrX 17433304 92 - 21770863 -----CCACAGCUAUCGCCGCGCCACAAGAUC-UGGCAACCCCCAAUGUUGCCAGCAGAUGCG---ACGCAGCAUGUUGCCAAC--------CAAAAGUAAGCGAGCGA -----.........(((((((...........-(((((((.....(((((((..((....)).---..))))))))))))))..--------.........))).)))) ( -30.05, z-score = -2.81, R) >droSec1.super_4 3548257 92 - 6179234 -----CCACAGCUAUUGCCGCGCCACAAGAUC-UGGCAACCCCAAAUGUUGCCAGCAGAUGCG---UGGCAGCAUGUUGCCAAC--------CGAAAGUAAACGAGCAA -----.....(((.(((((((((........(-(((((((.......)))))))).....)))---))))))..((((......--------(....)..))))))).. ( -30.32, z-score = -2.18, R) >droSim1.chrX 3063921 92 + 17042790 -----CCACAGCUAUUGCCGCGCCACAAGAUC-UGGCAACCCCAAAUGUUGCCAGCAGAUGCG---UGGCAGCAUGUUGCCAAC--------CGAAAGUAAACGAGCAA -----.....(((.(((((((((........(-(((((((.......)))))))).....)))---))))))..((((......--------(....)..))))))).. ( -30.32, z-score = -2.18, R) >droWil1.scaffold_181150 2642132 96 - 4952429 -------GAAAUGGAAAUGCCGCCAGAUGAUGAUGAUGACCGGAGACGACGUUCGAAAAUGUU---GCUAAGCAUGUUGCCAGCGC---UCACAGCAGUAAACCGACAA -------....(((........)))...............(((((.(((((((....))))))---)))..((.(((.((....))---..)))))......))).... ( -15.40, z-score = 1.93, R) >consensus _____CCACAGCUAUUGCCGCGCCACAAGAUC_UGGCAACCCCAAAUGUUGCCAGCAGAUGCG___ACGCAGCAUGUUGCCAAC________CAAAAGUAAACGAGCAA ..........((....))...((..........(((((((.......)))))))((((((((.........)))).)))).........................)).. (-11.21 = -11.09 + -0.12)
| Location | 4,100,155 – 4,100,247 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 65.70 |
| Shannon entropy | 0.63035 |
| G+C content | 0.55347 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -16.04 |
| Energy contribution | -15.11 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980770 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
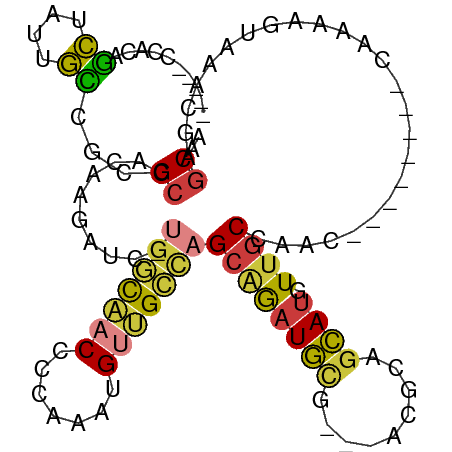
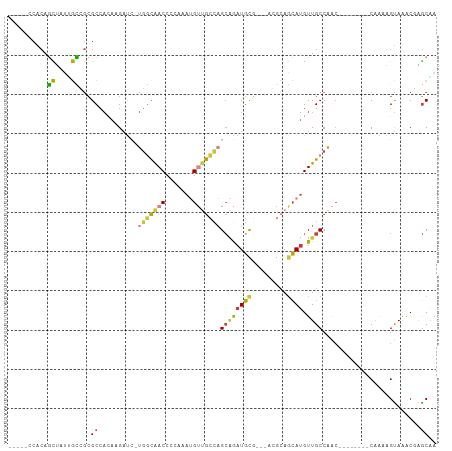
>dm3.chrX 4100155 92 - 22422827 UUGCUUGAUUACUUUCG--------GUUGGCAACAUGCUGCGU---CGCAUCGGCUGGCAACAUUUGGGGUUGCCA-GAUCUUGUGGCGCGGCAAUAGCUGUGG----- ...............((--------((((....).((((((((---((((....((((((((.......)))))))-)....))))))))))))..)))))...----- ( -39.90, z-score = -3.40, R) >droAna3.scaffold_13117 273503 109 + 5790199 UUGCCCGCCUGCCUCUGCAACAUGCCUCGGCAACCUUGUUGCCGCGAAAAUCCGCUCUCGCCGCUUCAUGUUGGUGCUGUCGCUGCGCUCACUUCACAGUGGGCCUCGG ..((((((.((......(((((((....((((((...))))))(((......)))...........)))))))((((.......))))........))))))))..... ( -35.50, z-score = -0.93, R) >droEre2.scaffold_4690 1470417 92 - 18748788 UCGCUGGUUUACUUUUG--------GUUGGCAACAUGCUGCGU---CGCAUCUGCUGGCAACAUUGGGGGUUGCCA-GAUCUUGUGGCGCGGCAAUAGCUGUGG----- ..((((((..((.....--------))..))....((((((((---((((....((((((((.......)))))))-)....)))))))))))).)))).....----- ( -41.50, z-score = -3.62, R) >droYak2.chrX 17433304 92 + 21770863 UCGCUCGCUUACUUUUG--------GUUGGCAACAUGCUGCGU---CGCAUCUGCUGGCAACAUUGGGGGUUGCCA-GAUCUUGUGGCGCGGCGAUAGCUGUGG----- ..(((.(((.((.....--------)).)))....((((((((---((((....((((((((.......)))))))-)....))))))))))))..))).....----- ( -41.50, z-score = -3.20, R) >droSec1.super_4 3548257 92 + 6179234 UUGCUCGUUUACUUUCG--------GUUGGCAACAUGCUGCCA---CGCAUCUGCUGGCAACAUUUGGGGUUGCCA-GAUCUUGUGGCGCGGCAAUAGCUGUGG----- ...............((--------((((....).((((((..---((((....((((((((.......)))))))-)....))))..))))))..)))))...----- ( -32.80, z-score = -0.92, R) >droSim1.chrX 3063921 92 - 17042790 UUGCUCGUUUACUUUCG--------GUUGGCAACAUGCUGCCA---CGCAUCUGCUGGCAACAUUUGGGGUUGCCA-GAUCUUGUGGCGCGGCAAUAGCUGUGG----- ...............((--------((((....).((((((..---((((....((((((((.......)))))))-)....))))..))))))..)))))...----- ( -32.80, z-score = -0.92, R) >droWil1.scaffold_181150 2642132 96 + 4952429 UUGUCGGUUUACUGCUGUGA---GCGCUGGCAACAUGCUUAGC---AACAUUUUCGAACGUCGUCUCCGGUCAUCAUCAUCAUCUGGCGGCAUUUCCAUUUC------- ...(((((((((....))))---))((((((.....)).))))---........))).....(((.((((.............)))).)))...........------- ( -21.72, z-score = -0.02, R) >consensus UUGCUCGUUUACUUUUG________GUUGGCAACAUGCUGCGU___CGCAUCUGCUGGCAACAUUUGGGGUUGCCA_GAUCUUGUGGCGCGGCAAUAGCUGUGG_____ .........................((((....)).))........((((.....(((((((.......)))))))......)))).((((((....))))))...... (-16.04 = -15.11 + -0.93)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:47 2011