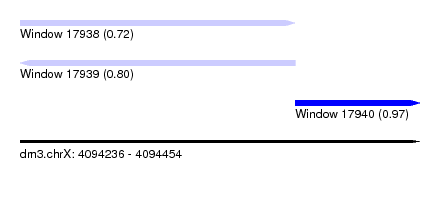
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,094,236 – 4,094,454 |
| Length | 218 |
| Max. P | 0.971249 |

| Location | 4,094,236 – 4,094,386 |
|---|---|
| Length | 150 |
| Sequences | 4 |
| Columns | 150 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Shannon entropy | 0.12943 |
| G+C content | 0.38938 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -30.43 |
| Energy contribution | -30.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.724204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
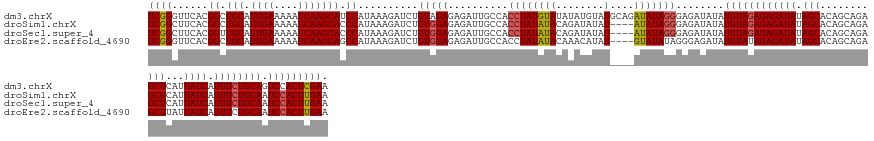
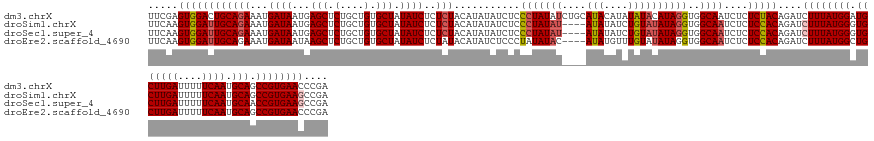
>dm3.chrX 4094236 150 + 22422827 UCGGGUUCACGGCUGCAUUGAAAAAUCAAGCAUCCAUAAAGAUCUGUAGAGAGAUUGCCACCUAUGUAUAUAUGUAUGCAGAUAUAGGGAGAUAUAUGUAGAGAGAUAUAGCACAGCAGAGCUCAUUAUCAUUUCUGCAGUCCACUCGAA ((((((..((((((((.((((....)))))))........(((((......)))))))).(((((((.....((....)).)))))))........((((((((((((.(((........)))...)))).))))))))))..)))))). ( -40.50, z-score = -1.40, R) >droSim1.chrX 3057895 146 + 17042790 UCGGCUUCACGGCUGCAUUGAAAAAUCAAGCACCCAUAAAGAUCUGUGGAGAGAUUGCCACCUAUAUACAGAUAUAU----AUAUAGGGAGAUAUAUGUAGAGAGAUAUAGCACAGCAGAGCUCAUUAUCAUUUCUGCAAUCCACUUGAA ..(((.....((.(((.((((....))))))).)).....(((((......)))))))).((((((((........)----)))))))(((.....((((((((((((.(((........)))...)))).)))))))).....)))... ( -37.80, z-score = -1.79, R) >droSec1.super_4 3542412 146 - 6179234 UCGGCUUCACGGUUGCAUUGAAAAAUCAAGCACCCAUAAAGAUCUGUGGAGAGAUUGCCACCUAUAUACAGAUAUAU----AUAUAGGGAGAUAUAUGUAGAGAGAUAUAGCACAGCAGAGCUCAUUAUCAUUUCUGCAAUCCACUUGAA ..(((.....((.(((.((((....))))))).)).....(((((......)))))))).((((((((........)----)))))))(((.....((((((((((((.(((........)))...)))).)))))))).....)))... ( -37.80, z-score = -2.01, R) >droEre2.scaffold_4690 1464450 146 + 18748788 UCGGGUUCACGGCUGCAUUGAAAAAUCAAGCAGCCAUAAAGAUCUGUGGAGAGAUUGCCACCUAUAUACAAACAUAU----GUAUAUAGGGAGAUAUGUAUAGAGAUAUAGCACAGCAGAGCUUAUUAUCAUUUCUGCAAUCCACUUGAA ((((((....((((((.((((....)))))))))).....(((.((..((((.....(..((((((((((......)----)))))))))..)...........((((.(((........)))...)))).))))..))))).)))))). ( -45.40, z-score = -3.90, R) >consensus UCGGCUUCACGGCUGCAUUGAAAAAUCAAGCACCCAUAAAGAUCUGUGGAGAGAUUGCCACCUAUAUACAGAUAUAU____AUAUAGGGAGAUAUAUGUAGAGAGAUAUAGCACAGCAGAGCUCAUUAUCAUUUCUGCAAUCCACUUGAA ((((......((.(((.((((....))))))).))..........(((((..........(((((((..............)))))))........((((((((((((.(((........)))...)))).)))))))).))))))))). (-30.43 = -30.86 + 0.44)
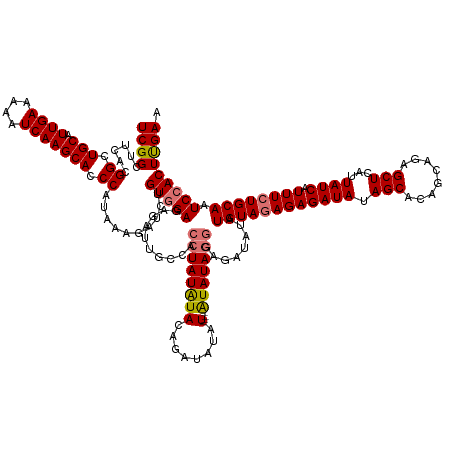
| Location | 4,094,236 – 4,094,386 |
|---|---|
| Length | 150 |
| Sequences | 4 |
| Columns | 150 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Shannon entropy | 0.12943 |
| G+C content | 0.38938 |
| Mean single sequence MFE | -41.75 |
| Consensus MFE | -30.49 |
| Energy contribution | -31.11 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.804971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
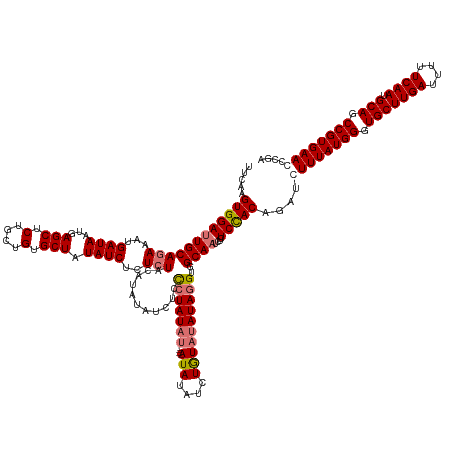
>dm3.chrX 4094236 150 - 22422827 UUCGAGUGGACUGCAGAAAUGAUAAUGAGCUCUGCUGUGCUAUAUCUCUCUACAUAUAUCUCCCUAUAUCUGCAUACAUAUAUACAUAGGUGGCAAUCUCUCUACAGAUCUUUAUGGAUGCUUGAUUUUUCAAUGCAGCCGUGAACCCGA ...(((.(((.((((((...((((...(((.(....).))).))))..))).....(((((...(((((.((....)).)))))...)))))))).))))))........(((((((.(((((((....)))).))).)))))))..... ( -32.70, z-score = 0.25, R) >droSim1.chrX 3057895 146 - 17042790 UUCAAGUGGAUUGCAGAAAUGAUAAUGAGCUCUGCUGUGCUAUAUCUCUCUACAUAUAUCUCCCUAUAU----AUAUAUCUGUAUAUAGGUGGCAAUCUCUCCACAGAUCUUUAUGGGUGCUUGAUUUUUCAAUGCAGCCGUGAAGCCGA .....((((((((((((...((((...(((.(....).))).))))..)))...........(((((((----(((....))))))))))..))))....)))))....((((((((.(((((((....)))).))).)))))))).... ( -44.60, z-score = -2.87, R) >droSec1.super_4 3542412 146 + 6179234 UUCAAGUGGAUUGCAGAAAUGAUAAUGAGCUCUGCUGUGCUAUAUCUCUCUACAUAUAUCUCCCUAUAU----AUAUAUCUGUAUAUAGGUGGCAAUCUCUCCACAGAUCUUUAUGGGUGCUUGAUUUUUCAAUGCAACCGUGAAGCCGA .....((((((((((((...((((...(((.(....).))).))))..)))...........(((((((----(((....))))))))))..))))....)))))....((((((((.(((((((....)))).))).)))))))).... ( -42.50, z-score = -2.83, R) >droEre2.scaffold_4690 1464450 146 - 18748788 UUCAAGUGGAUUGCAGAAAUGAUAAUAAGCUCUGCUGUGCUAUAUCUCUAUACAUAUCUCCCUAUAUAC----AUAUGUUUGUAUAUAGGUGGCAAUCUCUCCACAGAUCUUUAUGGCUGCUUGAUUUUUCAAUGCAGCCGUGAACCCGA .....((((((((((((.((((((((((((.(....).))).)))...))).))).))).(((((((((----(......))))))))))..))))....))))).....(((((((((((((((....)))).)))))))))))..... ( -47.20, z-score = -4.54, R) >consensus UUCAAGUGGAUUGCAGAAAUGAUAAUGAGCUCUGCUGUGCUAUAUCUCUCUACAUAUAUCUCCCUAUAU____AUAUAUCUGUAUAUAGGUGGCAAUCUCUCCACAGAUCUUUAUGGGUGCUUGAUUUUUCAAUGCAGCCGUGAACCCGA .....(((((..(((((.............)))))..((((((..........((((((..............))))))..........)))))).....)))))....((((((((.(((((((....)))).))).)))))))).... (-30.49 = -31.11 + 0.62)
| Location | 4,094,386 – 4,094,454 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Shannon entropy | 0.33386 |
| G+C content | 0.37360 |
| Mean single sequence MFE | -14.38 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.42 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
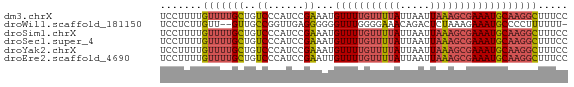
>dm3.chrX 4094386 68 + 22422827 UCCUUUUGUUUUGCUGUCCCAUCCGAAAUGUUUUGUUUUAUUAAUUAAAGCGAAAUGCAAGGCUUUCC .......(((((((..((......))...(((((((((((.....))))))))))))))))))..... ( -14.20, z-score = -2.85, R) >droWil1.scaffold_181150 2634384 65 - 4952429 UCCUCUUGUU--GUUGCCGGUUGAGGGGGGUUUGGGGAAACAGACUCUAAAGAAAUGCCCCUUUUUU- .((((.....--..........))))(((((((((((.......))))))......)))))......- ( -15.26, z-score = 1.21, R) >droSim1.chrX 3058041 68 + 17042790 UCCUUUUGUUUUGCUGUCCCAUCCGAAAUGUUUUGUUUUAUUAAUUAAAGCGAAAUGCAAGGCUUUCC .......(((((((..((......))...(((((((((((.....))))))))))))))))))..... ( -14.20, z-score = -2.85, R) >droSec1.super_4 3542558 68 - 6179234 UCCUUUUGUUUUGCUGUCCCAUCCGAAAUGUUUUGUUUUAUUAAUUAAAGCGAAAUGCAAGGCUUUCC .......(((((((..((......))...(((((((((((.....))))))))))))))))))..... ( -14.20, z-score = -2.85, R) >droYak2.chrX 17427758 68 - 21770863 UCCUUUUGUUUUGCUGUCCCAUCCGAAAUGUUUUGUUUUAUUAAUUAAAGCGAAAUGCAAGGCUUUCC .......(((((((..((......))...(((((((((((.....))))))))))))))))))..... ( -14.20, z-score = -2.85, R) >droEre2.scaffold_4690 1464596 68 + 18748788 UCCUUUUGUUUUGCUGUCCCAUCCGAAUUGUUUUGUUUUAUUAAUUAAAGCGAAAUGCAAGGCUUUCC .......(((((((..((......))...(((((((((((.....))))))))))))))))))..... ( -14.20, z-score = -2.93, R) >consensus UCCUUUUGUUUUGCUGUCCCAUCCGAAAUGUUUUGUUUUAUUAAUUAAAGCGAAAUGCAAGGCUUUCC .......(((((((..((......))...(((((((((((.....))))))))))))))))))..... (-10.75 = -11.42 + 0.67)
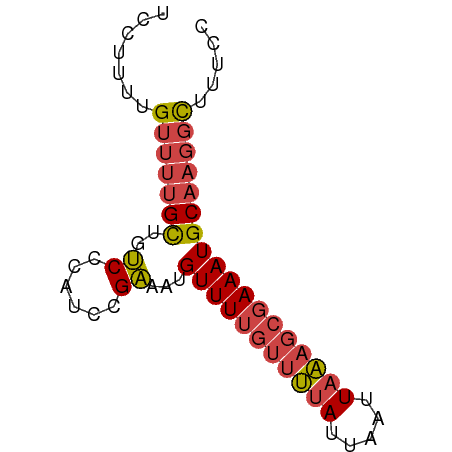
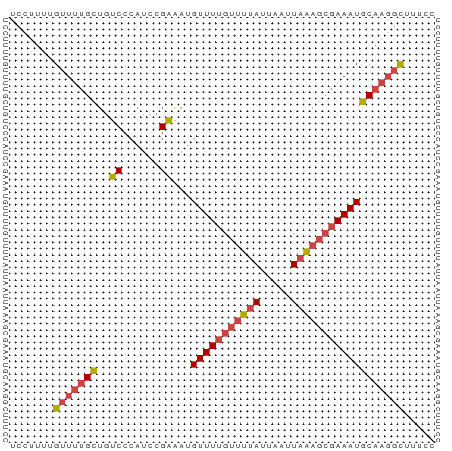
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:44 2011