| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,051,260 – 4,051,369 |
| Length | 109 |
| Max. P | 0.868541 |

| Location | 4,051,260 – 4,051,369 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.69 |
| Shannon entropy | 0.52820 |
| G+C content | 0.35670 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -12.25 |
| Energy contribution | -13.92 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.49 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4051260 109 + 22422827 UGGAGUGUGUUGAUAGCGAUUAAUGGAUUUACUUGUUUAUUGAUGUUUUUAUUUAACACCCAUAAAUGUGAGUAUUCAGUUUGGCUUAAUGCCAAGCGAUAGCGCAUUG----------- (((.((((..(((.(((.((((((((((......))))))))))))).)))....)))))))..((((((..((((..(((((((.....))))))))))).)))))).----------- ( -32.60, z-score = -3.73, R) >droEre2.scaffold_4690 1428291 106 + 18748788 UGGAGUGUGUUGAUAGCGAUUAAUGGAUUUACUUGUUUAUUGAUGUUUUUAUUUAACACCAAUAAAUGUGGG--UUCAGUUUGGCUAGAUGCUA-CCGAAAGCGCAUUG----------- ...((((((((.....((.....(((((((((..((((((((.((((.......)))).)))))))))))))--))))...((((.....))))-.))..)))))))).----------- ( -27.20, z-score = -2.09, R) >droYak2.chrX 17392833 86 - 21770863 UGGAGUGUGUUGAUAGCGAUUAAUG--------------------UUUUAAUUUAACACCGAUAAAUGUGGA--UUCAGUUUGGCUUAAUGGUA-CCGAAAACGCAUUG----------- ....(.(((((((.((((.....))--------------------)).....))))))))....((((((..--(((.((...(((....))))-).)))..)))))).----------- ( -14.90, z-score = 0.37, R) >droSec1.super_4 3507615 108 - 6179234 UGGAGUGUGUUGAUAGCGAUUAAUGGAUUUACUUGUUUAUUGAUGUUUUUAUUUAACACCCAUAAAUGCGAGUGUUCAGUUUGGCUUAAUGCUA-GCGAAAGCGCAUUG----------- (((.((((..(((.(((.((((((((((......))))))))))))).)))....)))))))..(((((.((((((.((.....)).)))))).-((....))))))).----------- ( -30.60, z-score = -2.79, R) >droSim1.chrX 3024885 108 + 17042790 UGGAGUGUGUUGAUAGCGAUUAAUGGAUUUACUUGUUUAUUGAUGUUUUUAUUUAACACCCAUAAAUGUGAGUGUUCAGUUUGGCUUAAUGCUA-GCGAAAGCGCAUUG----------- (((.((((..(((.(((.((((((((((......))))))))))))).)))....)))))))..(((((.((((((.((.....)).)))))).-((....))))))).----------- ( -28.20, z-score = -2.25, R) >droWil1.scaffold_181150 2596074 114 - 4952429 CGAGUCUUGUCUUAAGUCAUUAAUUAAC-----UGAUUGCUUAUGCAUUUAUAGAGCAACCAAAAGUAUGCA-ACACUCUUUGUUUUGAUGGAAAACUGCUGUUGGUUGUCCUUGCAAUG ............(((((.((((......-----)))).)))))((((.....((..(((((((.((((....-.((.((........))))......)))).)))))))..))))))... ( -22.10, z-score = 0.07, R) >consensus UGGAGUGUGUUGAUAGCGAUUAAUGGAUUUACUUGUUUAUUGAUGUUUUUAUUUAACACCCAUAAAUGUGAG__UUCAGUUUGGCUUAAUGCUA_GCGAAAGCGCAUUG___________ ......(((((((.(((.((((((((((......))))))))))))).....))))))).....((((((....(((.((.((((.....)))).)))))..))))))............ (-12.25 = -13.92 + 1.67)

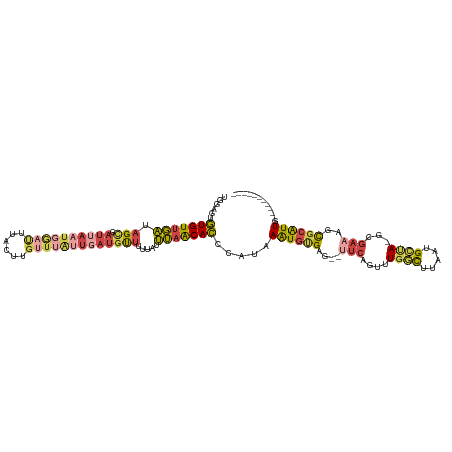

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:36 2011