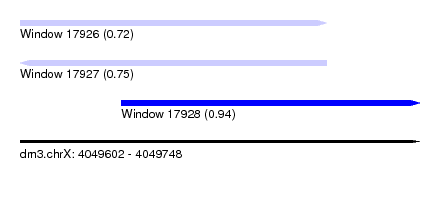
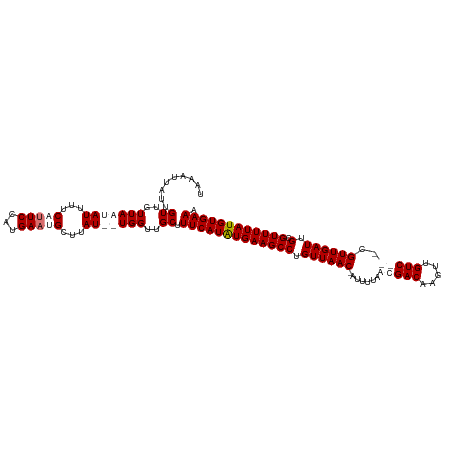
| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,049,602 – 4,049,748 |
| Length | 146 |
| Max. P | 0.944342 |

| Location | 4,049,602 – 4,049,714 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Shannon entropy | 0.12441 |
| G+C content | 0.30566 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -18.80 |
| Energy contribution | -21.30 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.720002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4049602 112 + 22422827 UAAAUUAUAGUUGUUAUUAUUUUCCUUCCAUGAAUGCUUAU--UGGUUGCUUUCAUGUGAAGCCUGUUAACGAUUUUAACGACAAAAUGUC---CGUUGAUUGCGUUUUAUGUGAAA ....(((((...............((((((((((.((....--.....)).)))))).))))......((((...((((((((.....)).---))))))...))))...))))).. ( -22.60, z-score = -1.95, R) >droSim1.chrX 3022803 116 + 17042790 UAAAUUAUUGUUGUUAAUAUUUUCAUUCCAUGAAUGCUUAUAUUGGUUGCUUUCAUAUGAAGCCUGUUAAC-AUUUUAACGACAAGUUGUCGUCCGUUGAUUGCGUUUUAUGUGAAA .........((..(((((((...(((((...)))))...)))))))..)).(((((((((((((.((((((-......(((((.....)))))..)))))).).)))))))))))). ( -28.40, z-score = -3.27, R) >droSec1.super_4 3506032 116 - 6179234 UAAAUUAUUGUUGUUAAUAUUUUCAUUCCAUGAAUGCUUAUAUUGGUUGCUUUCAUAUGAAGCCUGUUAAC-AUUUUAACGACAAGUUGUCGUUCGUUGAUUGCGUUUUAUGUGAAA .........((..(((((((...(((((...)))))...)))))))..)).(((((((((((((.((((((-.....((((((.....)))))).)))))).).)))))))))))). ( -29.60, z-score = -3.61, R) >droEre2.scaffold_4690 1426598 111 + 18748788 UAAAUUAUUGUUGUUAUUAUAUUCUGUCCAUGAAUGCUUAU--UGGUUGCUUUCAUGUGAAGCCUGUUAAC-AUUUUAACGACAACUUGUC---CGUUGAUUGCGUUUUAUGUGAAA ...........(((((..(((..((.((((((((.((....--.....)).)))))).))))..)))))))-)..((((((((.....)).---))))))(..(((...)))..).. ( -17.60, z-score = 0.01, R) >consensus UAAAUUAUUGUUGUUAAUAUUUUCAUUCCAUGAAUGCUUAU__UGGUUGCUUUCAUAUGAAGCCUGUUAAC_AUUUUAACGACAAGUUGUC___CGUUGAUUGCGUUUUAUGUGAAA .........((..(((((((...(((((...)))))...)))))))..)).(((((((((((((.((((((.......(((((.....)))))..)))))).).)))))))))))). (-18.80 = -21.30 + 2.50)

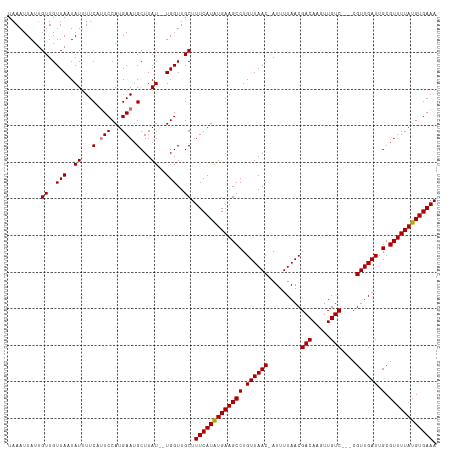
| Location | 4,049,602 – 4,049,714 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Shannon entropy | 0.12441 |
| G+C content | 0.30566 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4049602 112 - 22422827 UUUCACAUAAAACGCAAUCAACG---GACAUUUUGUCGUUAAAAUCGUUAACAGGCUUCACAUGAAAGCAACCA--AUAAGCAUUCAUGGAAGGAAAAUAAUAACAACUAUAAUUUA ((((......((((.....((((---.((.....)))))).....))))......((((.((((((.((.....--....)).)))))))))))))).................... ( -19.30, z-score = -2.73, R) >droSim1.chrX 3022803 116 - 17042790 UUUCACAUAAAACGCAAUCAACGGACGACAACUUGUCGUUAAAAU-GUUAACAGGCUUCAUAUGAAAGCAACCAAUAUAAGCAUUCAUGGAAUGAAAAUAUUAACAACAAUAAUUUA (((((........((....((((((((((.....))))))....)-))).....))(((.((((((.((...........)).)))))))))))))).................... ( -17.80, z-score = -1.59, R) >droSec1.super_4 3506032 116 + 6179234 UUUCACAUAAAACGCAAUCAACGAACGACAACUUGUCGUUAAAAU-GUUAACAGGCUUCAUAUGAAAGCAACCAAUAUAAGCAUUCAUGGAAUGAAAAUAUUAACAACAAUAAUUUA (((((........((....((((((((((.....))))))....)-))).....))(((.((((((.((...........)).)))))))))))))).................... ( -17.40, z-score = -1.78, R) >droEre2.scaffold_4690 1426598 111 - 18748788 UUUCACAUAAAACGCAAUCAACG---GACAAGUUGUCGUUAAAAU-GUUAACAGGCUUCACAUGAAAGCAACCA--AUAAGCAUUCAUGGACAGAAUAUAAUAACAACAAUAAUUUA ..........((((....((((.---.....)))).))))....(-((((.....((((.((((((.((.....--....)).)))))))).)).......)))))........... ( -16.30, z-score = -1.47, R) >consensus UUUCACAUAAAACGCAAUCAACG___GACAACUUGUCGUUAAAAU_GUUAACAGGCUUCACAUGAAAGCAACCA__AUAAGCAUUCAUGGAAUGAAAAUAAUAACAACAAUAAUUUA ((((.........((...........(((.....)))(((((.....)))))..))(((.((((((.((...........)).))))))))).)))).................... (-14.93 = -15.17 + 0.25)

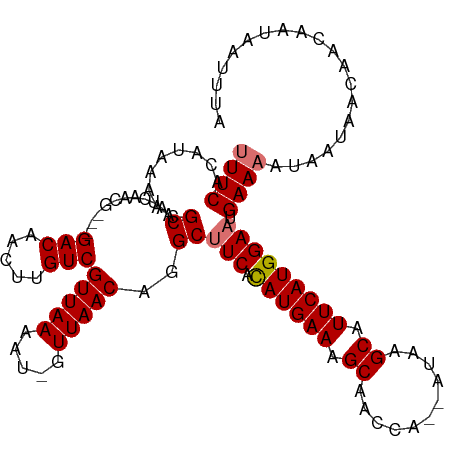
| Location | 4,049,639 – 4,049,748 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Shannon entropy | 0.12518 |
| G+C content | 0.30227 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.14 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4049639 109 + 22422827 --UUAUUGGUUGCUUUCAUGUGAAGCCUGUUAACGAUUUUAACGACAAAAUGUC---CGUUGAUUGCGUUUUAUGUGAAAUACUGUAACAAUUAAAAAUCAUCAAUUUUCCUCU --......(((((((((((((((((((.(((((((........(((.....)))---))))))).).)))))))))))))....)))))......................... ( -23.00, z-score = -2.43, R) >droSim1.chrX 3022840 113 + 17042790 UUAUAUUGGUUGCUUUCAUAUGAAGCCUGUUAAC-AUUUUAACGACAAGUUGUCGUCCGUUGAUUGCGUUUUAUGUGAAAUACUGUAACAAUUAAAAAUCAUCAAUUUUCCUCU ........(((((((((((((((((((.((((((-......(((((.....)))))..)))))).).)))))))))))))....)))))......................... ( -25.90, z-score = -3.20, R) >droSec1.super_4 3506069 113 - 6179234 UUAUAUUGGUUGCUUUCAUAUGAAGCCUGUUAAC-AUUUUAACGACAAGUUGUCGUUCGUUGAUUGCGUUUUAUGUGAAAUACUGUAACAAUUAAAAAUCAUCAAUUUUCCUCU ........(((((((((((((((((((.((((((-.....((((((.....)))))).)))))).).)))))))))))))....)))))......................... ( -27.10, z-score = -3.53, R) >droYak2.chrX 17391207 107 - 21770863 --UUAUUGGUUGCUUUCAUGUGAAGCUUGUUAAC-AUUUUAACGACAACUUGUC---CGUUGAUUGCAUUUUAUGUGAAAUACUGUAACAAUUAAAAAUCAUUA-UUUUCCUCU --......(((((((((((((((((((((((((.-...)))))))((((.....---.))))...))..)))))))))))....)))))...............-......... ( -18.40, z-score = -1.22, R) >droEre2.scaffold_4690 1426635 107 + 18748788 --UUAUUGGUUGCUUUCAUGUGAAGCCUGUUAAC-AUUUUAACGACAACUUGUC---CGUUGAUUGCGUUUUAUGUGAAAUAAUGUAACAAUUAAAAAUCAUUA-UUUUCCUCU --......((((((((((((((((((.((((((.-...)))))).((((.....---.)))).....)))))))))))))....)))))...............-......... ( -20.30, z-score = -1.85, R) >consensus __UUAUUGGUUGCUUUCAUGUGAAGCCUGUUAAC_AUUUUAACGACAACUUGUC___CGUUGAUUGCGUUUUAUGUGAAAUACUGUAACAAUUAAAAAUCAUCAAUUUUCCUCU ........(((((((((((((((((((.((((((.........(((.....)))....)))))).).)))))))))))))....)))))......................... (-19.34 = -19.14 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:34 2011