| Sequence ID | dm3.chrX |
|---|---|
| Location | 4,002,492 – 4,002,607 |
| Length | 115 |
| Max. P | 0.654505 |

| Location | 4,002,492 – 4,002,607 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 131 |
| Reading direction | reverse |
| Mean pairwise identity | 67.92 |
| Shannon entropy | 0.61432 |
| G+C content | 0.43929 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -10.42 |
| Energy contribution | -11.31 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.654505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 4002492 115 - 22422827 GCGAAUCUAUAUAU-UUAUUUAUUUGCACAUUUA-UGGCCAUAAAUGGUCGGAAAGCUGCGCCU------CAAG---CGGCUG--GCCAAUCACGUUGAUUGACUAACUGACUAACGGCCGCCGAUUU--- ((((((..(((...-.)))..)))))).......-.((((.....(((((((..((((((....------...)---)))))(--(.(((((.....))))).))..)))))))..))))........--- ( -35.60, z-score = -2.81, R) >droSim1.chrX 2974951 113 - 17042790 GCGAAUCU--AUAU-UUAUUUAUUUGCACAUUUA-UGGCCAUAAAUGGUCGGAAAGCUGCGCCU------CAAG---CGGCUG--GCCAAUCACGUUGAUUGACUAACUGACUAACGGCCGACGAUUU--- ((((((..--((..-..))..)))))).......-(((((.....(((((((..((((((....------...)---)))))(--(.(((((.....))))).))..)))))))..))))).......--- ( -34.00, z-score = -2.81, R) >droSec1.super_4 3459892 113 + 6179234 GCGAAUCU--AUAU-UUAUUUAUUUGCACAUUUA-UGGCCAUAAAUGGUCGGAAAGCUGCGCCU------CAAG---CGGCUG--GCCAAUCACGUUGAUUGACUAACUGACUAACGGCCGCCGAUUU--- ((((((..--((..-..))..)))))).......-.((((.....(((((((..((((((....------...)---)))))(--(.(((((.....))))).))..)))))))..))))........--- ( -33.70, z-score = -2.24, R) >droYak2.chrX 17344473 113 + 21770863 GCGAAUCU--AUAU-UUAUUUAUUUGCACAUUUA-UGGCCAUAAAUGGUCGGAAAGCUGCGCCU------CAAG---CGGCUG--GCCAAUCACGUUGAUUGACUAACUGACUAACGGCCGCCGAUUU--- ((((((..--((..-..))..)))))).......-.((((.....(((((((..((((((....------...)---)))))(--(.(((((.....))))).))..)))))))..))))........--- ( -33.70, z-score = -2.24, R) >droEre2.scaffold_4690 1379290 109 - 18748788 GCGAAUCU--AUAU-UUAUUUAUUUGCACAUUUA-UGGCCAUAAAUGGUCGGAAAGCUGCGCCU------CAAG---CGGCUG--GCCAAUCACGUUGAUUG----ACUGACUAACGGCCGCCGAUUU--- ((((((..--((..-..))..)))))).((((((-(....)))))))(((((..((((((....------...)---)))))(--(((..(((.(((....)----))))).....)))).)))))..--- ( -33.10, z-score = -1.93, R) >droWil1.scaffold_181150 2536578 121 + 4952429 AUGAAUCU--AUAU-UUAUACAUACACA---AUA-UGACCAUAAAUGGUUAUUUAGCUAAACUCUCGGGGCAAGCCUCGAUUGCUGCCAAUCACGAAGAUACAAUAUUCGAAUGACGUCGAGAGACUU--- ........--....-.............---..(-((((((....))))))).........(((((((((((.((.......)))))).....((((.........)))).......)))))))....--- ( -25.00, z-score = -1.49, R) >droGri2.scaffold_14853 2916599 112 + 10151454 -UAAAUCUACGCUUAACAGUCGUCUGUAU-UUUAUUGACCAUAUUUGGGUAUUAAGGCGCGC--------CGAUC--UUGCU---GAUAGUU-UGAUGAUUGACUUGACUACUAGCGGCGGCCGUUUA--- -......(((.....((((....))))..-........(((....))))))...(((((.((--------((...--.((((---(.(((((-.............))))).))))).))))))))).--- ( -25.62, z-score = 0.06, R) >droMoj3.scaffold_6328 2313490 122 - 4453435 -UAAAUCUACGCUUAAUAGUCGUCUGUGUUUUUAUUGACCAUAUAUGGACAUAUUAAGGCGCGC------CUAAACGCAAUUC--GCCCCUGGCCGAUAGACGCUGAUUGACUAACGGCUGUCGGCCGUCG -.........((((((((...((((((((.............)))))))).)))))).))(((.------.....))).....--......(((((((((.((.((......)).)).))))))))).... ( -32.62, z-score = -0.76, R) >consensus GCGAAUCU__AUAU_UUAUUUAUUUGCACAUUUA_UGGCCAUAAAUGGUCGGAAAGCUGCGCCU______CAAG___CGGCUG__GCCAAUCACGUUGAUUGACUAACUGACUAACGGCCGCCGAUUU___ ..(((((..................(((.......((((((....))))))......))).................((((((....(((((.....))))).............))))))..)))))... (-10.42 = -11.31 + 0.89)
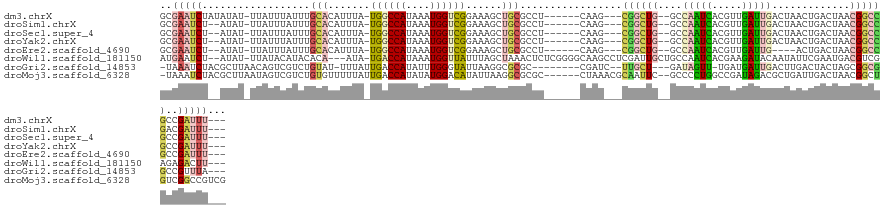


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:27 2011