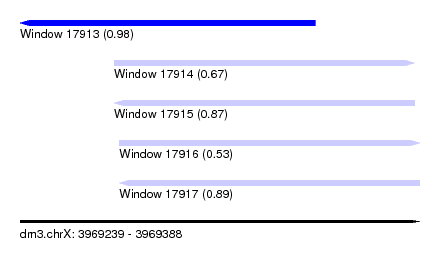
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,969,239 – 3,969,388 |
| Length | 149 |
| Max. P | 0.983096 |

| Location | 3,969,239 – 3,969,349 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 66.38 |
| Shannon entropy | 0.60624 |
| G+C content | 0.52307 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
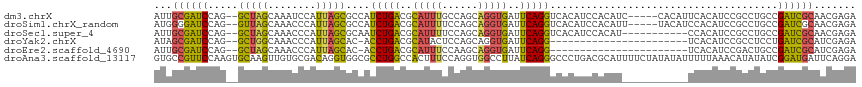
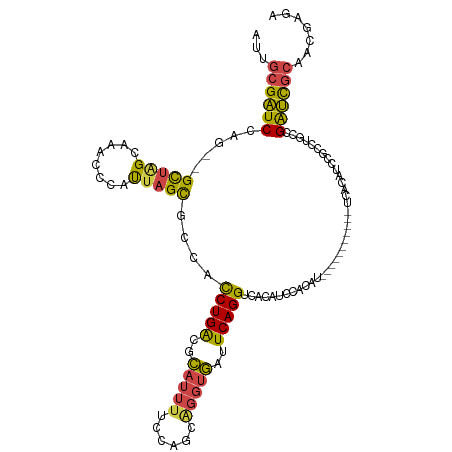
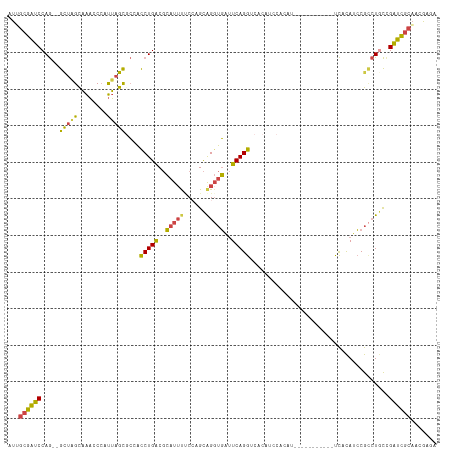
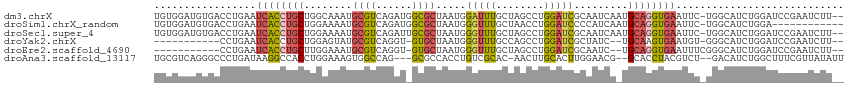
>dm3.chrX 3969239 110 - 22422827 AUUGCGAUCCAG--GCUAGCAAAUCCAUUAGCGCCAUCUGACGCAUUUGCCAGCAGGUGAUUCAGGUCACAUCCACAUC-----CACAUUCACAUCCGCCUGCCGAUCGCAACGAGA .(((((((((((--((..((..........))(..((((((..((((((....))))))..))))))..).........-----.............)))))..))))))))..... ( -32.00, z-score = -2.26, R) >droSim1.chrX_random 1547890 110 - 5698898 AUGGGGAUCCAG--GUUAGCAAACCCAUUAGCGCCAUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCACAUU-----UACAUCCACAUCCGCCUGCCGAUCGCAACGAGA ...(.((((..(--(((....)))).....(((.(....).)))........(((((((.....((.............-----.....)).....))))))).)))).)....... ( -22.17, z-score = 0.95, R) >droSec1.super_4 3426410 104 + 6179234 AUUGCGAUCCAG--GCUAGCAAACCCAUUAGCGCAAUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCACAU-----------CCACAUCCGCCUGCCGAUCGCAACGAGA .(((((((((((--((..((..........))(..((((((..(((((......)))))..))))))..)........-----------........)))))..))))))))..... ( -28.30, z-score = -1.88, R) >droYak2.chrX 17310453 91 + 21770863 AUAGCGAUCCAG--GCUGGCAAACCCAUUAGCAC-ACCUGACGCAUACUCCAGCAGGUGAUUCAGG-----------------------UCACAUCCGCCUCCUGAUCGCAUCGAGA ...((((((.((--(..(((.............(-(((((.(..........))))))).....((-----------------------......))))).)))))))))....... ( -29.90, z-score = -2.55, R) >droEre2.scaffold_4690 1344063 91 - 18748788 AUUGCGAUCCAG--GCUAGCAAACCCAUUAGCAC-ACCUGACGCAUUUCCAAGCAGGUGAUUCAGG-----------------------UCACAUCCGACUGCCGAUCGCAUCGAGA ..(((((((..(--((.................(-(((((.(..........))))))).....((-----------------------((......))))))))))))))...... ( -26.60, z-score = -2.11, R) >droAna3.scaffold_13117 134229 117 + 5790199 GUGCCGUUCCAAGUGCAAGUUGUGCGACAGGUGGCGCCUGGCCACUUUCCAGGUGGCCUUAUCAGGGCCCUGACGCAUUUUCUAUAUAUUUUUAAACAUAUAUCGGAUGAUUCAGGA ..((((..((...((((.....))))...))))))(((((((((((.....)))))))......))))(((((..(((((...((((((........)))))).)))))..))))). ( -37.40, z-score = -1.21, R) >consensus AUUGCGAUCCAG__GCUAGCAAACCCAUUAGCGCCACCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCACAU___________UCACAUCCGCCUGCCGAUCGCAACGAGA ...((((((.....(((((........)))))....(((((..(((((......)))))..)))))......................................))))))....... (-16.92 = -16.40 + -0.52)
| Location | 3,969,274 – 3,969,386 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.22 |
| Shannon entropy | 0.60443 |
| G+C content | 0.50990 |
| Mean single sequence MFE | -35.42 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.28 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.671066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
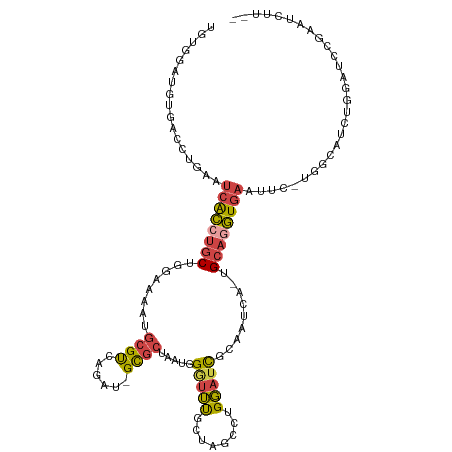
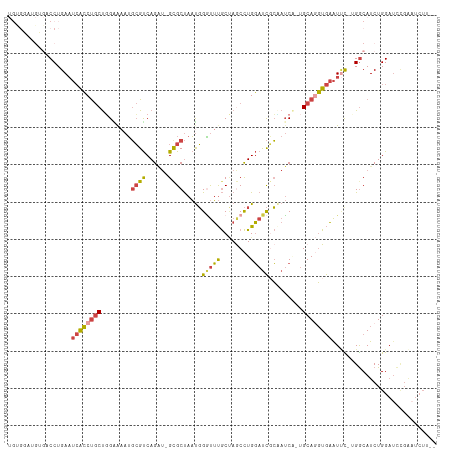
>dm3.chrX 3969274 112 + 22422827 UGUGGAUGUGACCUGAAUCACCUGCUGGCAAAUGCGUCAGAUGGCGCUAAUGGAUUUGCUAGCCUGGAUCGCAAUCAAUGCAGGUGAAUUC-UGGCAUCUGGAUCCGAAUCUU-- ..((((((((.((.((((((((((((((((((((((((....)))))......)))))))))).......(((.....)))))))).))))-.)))))....)))))......-- ( -39.80, z-score = -2.14, R) >droSim1.chrX_random 1547925 102 + 5698898 UGUGGAUGUGACCUGAAUCACCUGCUGGAAAAUGCGUCAGAUGGCGCUAAUGGGUUUGCUAACCUGGAUCCCCAUCAAUGCAGGUGAAUUC-UGGCAUCUGGA------------ ...(((((...((.(((((((((((..((....(((((....)))))...((((..(.(......).)..))))))...))))))).))))-.)))))))...------------ ( -38.00, z-score = -2.78, R) >droSec1.super_4 3426439 112 - 6179234 UGUGGAUGUGACCUGAAUCACCUGCUGGAAAAUGCGUCAGAUUGCGCUAAUGGGUUUGCUAGCCUGGAUCGCAAUCAAUGCAGGUGAAUUC-UGGCAUCUGGAUCCGAAUCUU-- ..((((((((.((.((((((((((((((........)))(((((((....((((((....))))))...)))))))...))))))).))))-.)))))....)))))......-- ( -38.90, z-score = -2.35, R) >droYak2.chrX 17310481 98 - 21770863 -----------CCUGAAUCACCUGCUGGAGUAUGCGUCAGGU-GUGCUAAUGGGUUUGCCAGCCUGGAUCGCUAUC--UGCAAGUGAAUGU-GGGCAUCUGGAUCCGAAUCUU-- -----------((((..((((.(((.(((....(((((((((-..((..........))..))))))..)))..))--)))).))))...)-)))..................-- ( -25.10, z-score = 1.36, R) >droEre2.scaffold_4690 1344091 99 + 18748788 -----------CCUGAAUCACCUGCUUGGAAAUGCGUCAGGU-GUGCUAAUGGGUUUGCUAGCCUGGAUCGCAAUC--UGCAGGUGAAUUUCGGGCAUCUGGAUCCGAAUCUU-- -----------((((((((((((((..(((..((((((((((-..((..........))..))))))..)))).))--)))))))))..))))))..................-- ( -34.70, z-score = -1.87, R) >droAna3.scaffold_13117 134269 107 - 5790199 UGCGUCAGGGCCCUGAUAAGGCCACCUGGAAAGUGGCCAG---GCGCCACCUGUCGCAC-AACUUGCACUUGGAACG--GCACCUACGUCU--GACAUCUGGCUUUCGUUAUAUU .(((((((....)))))..((((((.(....)))))))..---))((((..(((((((.-....)))....(((.((--.......)))))--))))..))))............ ( -36.00, z-score = -0.85, R) >consensus UGUGGAUGUGACCUGAAUCACCUGCUGGAAAAUGCGUCAGAU_GCGCUAAUGGGUUUGCUAGCCUGGAUCGCAAUCA_UGCAGGUGAAUUC_UGGCAUCUGGAUCCGAAUCUU__ .................((((((((........((((......)))).....(((((........))))).........))))))))............................ (-14.06 = -14.28 + 0.23)
| Location | 3,969,274 – 3,969,386 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.22 |
| Shannon entropy | 0.60443 |
| G+C content | 0.50990 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -13.56 |
| Energy contribution | -13.81 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
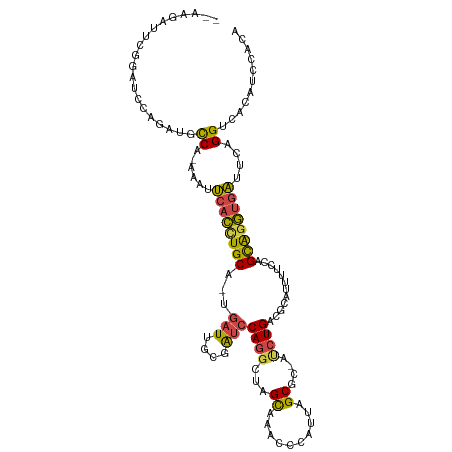
>dm3.chrX 3969274 112 - 22422827 --AAGAUUCGGAUCCAGAUGCCA-GAAUUCACCUGCAUUGAUUGCGAUCCAGGCUAGCAAAUCCAUUAGCGCCAUCUGACGCAUUUGCCAGCAGGUGAUUCAGGUCACAUCCACA --.......((((......(((.-((((.((((((((.....))).......(((.((((((.(.((((......)))).).)))))).)))))))))))).)))...))))... ( -31.30, z-score = -1.15, R) >droSim1.chrX_random 1547925 102 - 5698898 ------------UCCAGAUGCCA-GAAUUCACCUGCAUUGAUGGGGAUCCAGGUUAGCAAACCCAUUAGCGCCAUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCACA ------------....((((((.-((((.(((((((..(((((((................)))))))(((.(....).)))........))))))))))).))...)))).... ( -32.59, z-score = -2.57, R) >droSec1.super_4 3426439 112 + 6179234 --AAGAUUCGGAUCCAGAUGCCA-GAAUUCACCUGCAUUGAUUGCGAUCCAGGCUAGCAAACCCAUUAGCGCAAUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCACA --.......((((......(((.-((((.(((((((...(((((((.....((........))......))))))).((.......))..))))))))))).)))...))))... ( -31.50, z-score = -1.96, R) >droYak2.chrX 17310481 98 + 21770863 --AAGAUUCGGAUCCAGAUGCCC-ACAUUCACUUGCA--GAUAGCGAUCCAGGCUGGCAAACCCAUUAGCAC-ACCUGACGCAUACUCCAGCAGGUGAUUCAGG----------- --...................((-....((((((((.--(((((((...((((.((((..........)).)-))))).))).)).))..))))))))....))----------- ( -22.40, z-score = -0.10, R) >droEre2.scaffold_4690 1344091 99 - 18748788 --AAGAUUCGGAUCCAGAUGCCCGAAAUUCACCUGCA--GAUUGCGAUCCAGGCUAGCAAACCCAUUAGCAC-ACCUGACGCAUUUCCAAGCAGGUGAUUCAGG----------- --....(((((..........)))))..((((((((.--((.((((...((((...((..........))..-.)))).))))..))...))))))))......----------- ( -28.60, z-score = -2.38, R) >droAna3.scaffold_13117 134269 107 + 5790199 AAUAUAACGAAAGCCAGAUGUC--AGACGUAGGUGC--CGUUCCAAGUGCAAGUU-GUGCGACAGGUGGCGC---CUGGCCACUUUCCAGGUGGCCUUAUCAGGGCCCUGACGCA .......(....)...(.((((--((....((((((--((..((...((((....-.))))...))))))))---))(((((((.....)))))))...........))))))). ( -38.50, z-score = -1.04, R) >consensus __AAGAUUCGGAUCCAGAUGCCA_AAAUUCACCUGCA_UGAUUGCGAUCCAGGCUAGCAAACCCAUUAGCGC_AUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCACA ............................((((((((.......(((...((((...((..........))....)))).)))........))))))))................. (-13.56 = -13.81 + 0.25)

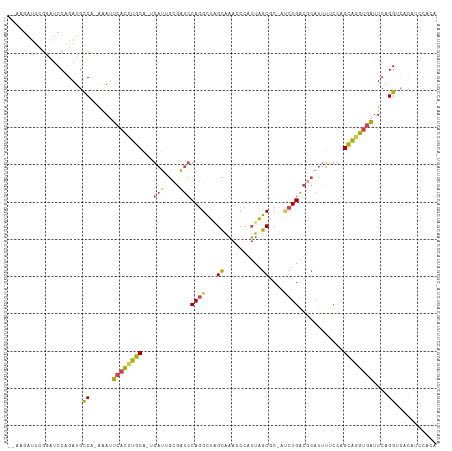
| Location | 3,969,276 – 3,969,388 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.58516 |
| G+C content | 0.50650 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -14.03 |
| Energy contribution | -14.10 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.525636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
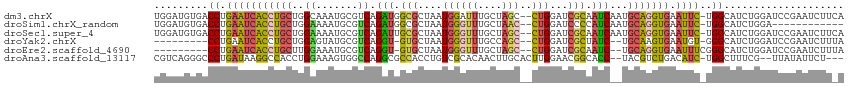
>dm3.chrX 3969276 112 + 22422827 UGGAUGUGACCUGAAUCACCUGCUGGCAAAUGCGUCAGAUGGCGCUAAUGGAUUUGCUAGC--CUGGAUCGCAAUCAAUGCAGGUGAAUUC-UGGCAUCUGGAUCCGAAUCUUCA .(((((((.((.((((((((((((((((((((((((....)))))......))))))))))--.......(((.....)))))))).))))-.)))))....))))......... ( -39.60, z-score = -1.96, R) >droSim1.chrX_random 1547927 100 + 5698898 UGGAUGUGACCUGAAUCACCUGCUGGAAAAUGCGUCAGAUGGCGCUAAUGGGUUUGCUAAC--CUGGAUCCCCAUCAAUGCAGGUGAAUUC-UGGCAUCUGGA------------ .(((((...((.(((((((((((..((....(((((....)))))...((((..(.(....--..).)..))))))...))))))).))))-.)))))))...------------ ( -38.00, z-score = -2.95, R) >droSec1.super_4 3426441 112 - 6179234 UGGAUGUGACCUGAAUCACCUGCUGGAAAAUGCGUCAGAUUGCGCUAAUGGGUUUGCUAGC--CUGGAUCGCAAUCAAUGCAGGUGAAUUC-UGGCAUCUGGAUCCGAAUCUUCA .(((((((.((.((((((((((((((........)))(((((((....((((((....)))--)))...)))))))...))))))).))))-.)))))....))))......... ( -38.70, z-score = -2.13, R) >droYak2.chrX 17310481 100 - 21770863 ---------CCUGAAUCACCUGCUGGAGUAUGCGUCAGGU-GUGCUAAUGGGUUUGCCAGC--CUGGAUCGCUAUC--UGCAAGUGAAUGU-GGGCAUCUGGAUCCGAAUCUUUA ---------((((..((((.(((.(((....(((((((((-..((..........))..))--))))..)))..))--)))).))))...)-))).................... ( -25.10, z-score = 1.43, R) >droEre2.scaffold_4690 1344091 101 + 18748788 ---------CCUGAAUCACCUGCUUGGAAAUGCGUCAGGU-GUGCUAAUGGGUUUGCUAGC--CUGGAUCGCAAUC--UGCAGGUGAAUUUCGGGCAUCUGGAUCCGAAUCUUUA ---------((((((((((((((..(((..((((((((((-..((..........))..))--))))..)))).))--)))))))))..)))))).................... ( -34.70, z-score = -1.88, R) >droAna3.scaffold_13117 134271 107 - 5790199 CGUCAGGGCCCUGAUAAGGCCACCUGGAAAGUGGCCAGGCGCCACCUGUCGCACAACUUGCACUUGGAACGGCACC--UACGUCUGACAUC-UGGCUUUCG--UUAUAUUCU--- .((((((.....(((..((((((.(....)))))))(((.(((.((.((.(((.....)))))..))...))).))--)..))).....))-)))).....--.........--- ( -34.30, z-score = -0.71, R) >consensus UGGAUGUGACCUGAAUCACCUGCUGGAAAAUGCGUCAGAUGGCGCUAAUGGGUUUGCUAGC__CUGGAUCGCAAUC__UGCAGGUGAAUUC_UGGCAUCUGGAUCCGAAUCUU_A ...............((((((((..((((((.(((.((......)).))).)))).((((...)))).......))...))))))))............................ (-14.03 = -14.10 + 0.07)
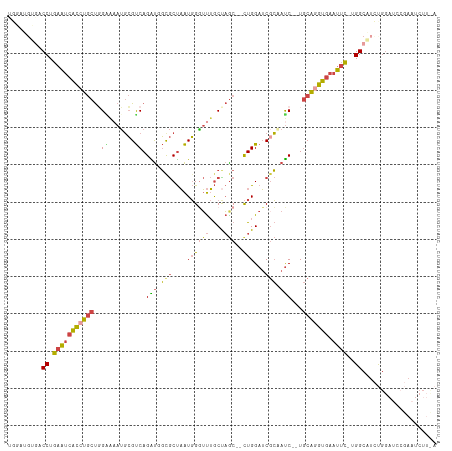
| Location | 3,969,276 – 3,969,388 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 68.74 |
| Shannon entropy | 0.58516 |
| G+C content | 0.50650 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -14.67 |
| Energy contribution | -14.43 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
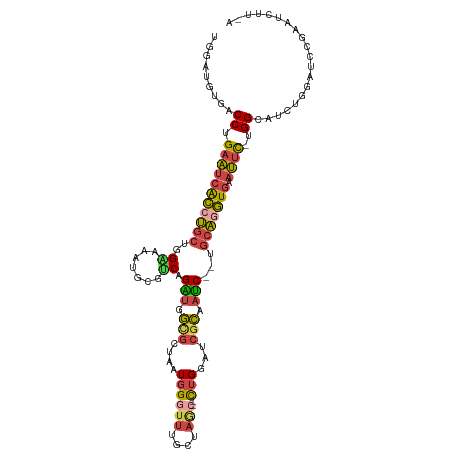
>dm3.chrX 3969276 112 - 22422827 UGAAGAUUCGGAUCCAGAUGCCA-GAAUUCACCUGCAUUGAUUGCGAUCCAG--GCUAGCAAAUCCAUUAGCGCCAUCUGACGCAUUUGCCAGCAGGUGAUUCAGGUCACAUCCA .........((((......(((.-((((.((((((((.....))).......--(((.((((((.(.((((......)))).).)))))).)))))))))))).)))...)))). ( -31.30, z-score = -0.73, R) >droSim1.chrX_random 1547927 100 - 5698898 ------------UCCAGAUGCCA-GAAUUCACCUGCAUUGAUGGGGAUCCAG--GUUAGCAAACCCAUUAGCGCCAUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCA ------------....((((((.-((((.(((((((..(((((((.......--.........)))))))(((.(....).)))........))))))))))).))...)))).. ( -32.59, z-score = -2.47, R) >droSec1.super_4 3426441 112 + 6179234 UGAAGAUUCGGAUCCAGAUGCCA-GAAUUCACCUGCAUUGAUUGCGAUCCAG--GCUAGCAAACCCAUUAGCGCAAUCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCA .........((((......(((.-((((.(((((((...(((((((.....(--(........))......))))))).((.......))..))))))))))).)))...)))). ( -31.50, z-score = -1.49, R) >droYak2.chrX 17310481 100 + 21770863 UAAAGAUUCGGAUCCAGAUGCCC-ACAUUCACUUGCA--GAUAGCGAUCCAG--GCUGGCAAACCCAUUAGCAC-ACCUGACGCAUACUCCAGCAGGUGAUUCAGG--------- .....................((-....((((((((.--(((((((...(((--(.((((..........)).)-))))).))).)).))..))))))))....))--------- ( -22.40, z-score = -0.19, R) >droEre2.scaffold_4690 1344091 101 - 18748788 UAAAGAUUCGGAUCCAGAUGCCCGAAAUUCACCUGCA--GAUUGCGAUCCAG--GCUAGCAAACCCAUUAGCAC-ACCUGACGCAUUUCCAAGCAGGUGAUUCAGG--------- ......(((((..........)))))..((((((((.--((.((((...(((--(...((..........))..-.)))).))))..))...))))))))......--------- ( -28.60, z-score = -2.58, R) >droAna3.scaffold_13117 134271 107 + 5790199 ---AGAAUAUAA--CGAAAGCCA-GAUGUCAGACGUA--GGUGCCGUUCCAAGUGCAAGUUGUGCGACAGGUGGCGCCUGGCCACUUUCCAGGUGGCCUUAUCAGGGCCCUGACG ---.........--(....)...-...(((((....(--(((((((..((...((((.....))))...))))))))))(((((((.....)))))))...........))))). ( -38.20, z-score = -1.35, R) >consensus U_AAGAUUCGGAUCCAGAUGCCA_GAAUUCACCUGCA__GAUUGCGAUCCAG__GCUAGCAAACCCAUUAGCGCCACCUGACGCAUUUUCCAGCAGGUGAUUCAGGUCACAUCCA ........................((((.(((((((...((.((((.((.....(((((........))))).......)))))).))....)))))))))))............ (-14.67 = -14.43 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:24 2011