| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,964,573 – 3,964,623 |
| Length | 50 |
| Max. P | 0.834351 |

| Location | 3,964,573 – 3,964,623 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 70.90 |
| Shannon entropy | 0.50863 |
| G+C content | 0.34939 |
| Mean single sequence MFE | -8.74 |
| Consensus MFE | -5.91 |
| Energy contribution | -5.69 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834351 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
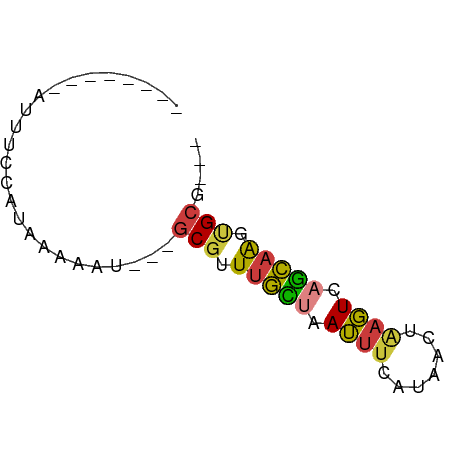
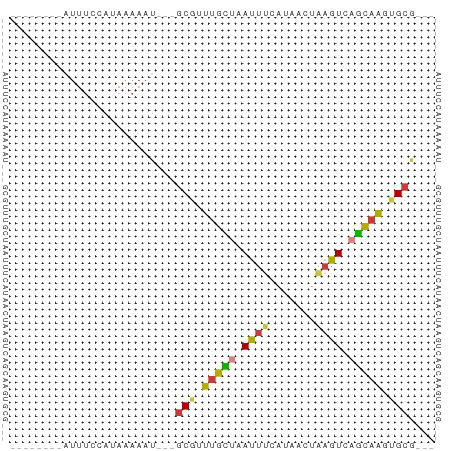
>dm3.chrX 3964573 50 + 22422827 --------AUUUCCAUAAAAAU---GCGUUUGUUAAUUUCAUAACUAAGUCAGCAAGUGCG--- --------..............---(((((((((.((((.......)))).))))))))).--- ( -7.20, z-score = -1.46, R) >droSim1.chrU 9394400 50 + 15797150 --------AUUUCCAUAAAAAU---GCGCUUGCUAAUUUCAUAACUAAGUCAGCAAAUGCA--- --------.............(---(((.(((((.((((.......)))).))))).))))--- ( -8.80, z-score = -1.39, R) >droSec1.super_4 3422089 50 - 6179234 --------AUUUCCAUAAAAAU---GCGCUUGCUAAUUUCAUAACUAAGUCAGCAAAUGCA--- --------.............(---(((.(((((.((((.......)))).))))).))))--- ( -8.80, z-score = -1.39, R) >droYak2.chrX 17305028 50 - 21770863 --------AUUUCCAUAAAAAU---GCGUUUGCUUAUUUCAUAACUAAGUCAGCCAGUGCG--- --------..............---(((((.(((.((((.......)))).))).))))).--- ( -5.50, z-score = -0.28, R) >droEre2.scaffold_4690 1335933 50 + 18748788 --------AUUUCCAUAAAAAU---GCGUUUGCUAAUUUCAUAACUAAGUCAGCAAGUGCG--- --------..............---(((((((((.((((.......)))).))))))))).--- ( -9.60, z-score = -2.20, R) >droVir3.scaffold_12928 6581491 56 + 7717345 AGUGCCAUGCAGAAAAAGAAAU---GCGUUUGCUAAUUUCAUAAAU--GUGCGCAGGGGCG--- .((.((.((((.(....(((((---((....))..))))).....)--.))))..)).)).--- ( -10.40, z-score = 0.94, R) >triCas2.chrUn_49 245918 56 - 463132 --------AUUUGCUUAAAAAUUCGGCGAUUUUUUAAUUCGCAAUUGAUUUUAAAGCCGGUCCC --------....((((.((((((..((((.........))))....)))))).))))....... ( -10.90, z-score = -1.14, R) >consensus ________AUUUCCAUAAAAAU___GCGUUUGCUAAUUUCAUAACUAAGUCAGCAAGUGCG___ .........................(((.(((((.((((.......)))).))))).))).... ( -5.91 = -5.69 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:20 2011