| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,953,811 – 3,953,901 |
| Length | 90 |
| Max. P | 0.933421 |

| Location | 3,953,811 – 3,953,901 |
|---|---|
| Length | 90 |
| Sequences | 11 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 75.09 |
| Shannon entropy | 0.50880 |
| G+C content | 0.43671 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -11.08 |
| Energy contribution | -11.49 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
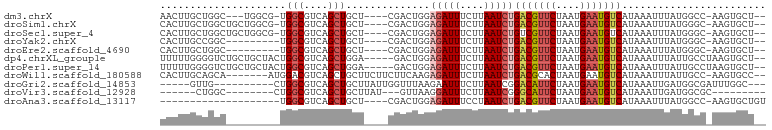
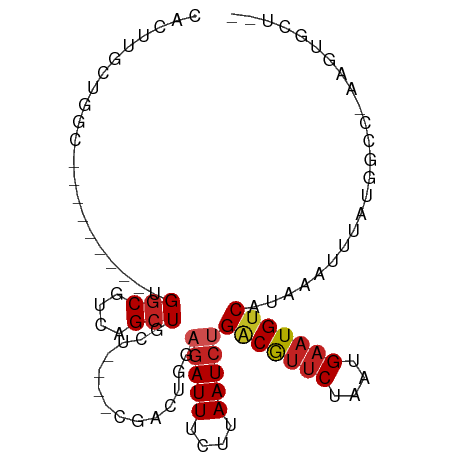
>dm3.chrX 3953811 90 + 22422827 AACUUGCUGGC---UGGCG-UGGCGUCAGCUGCU----CGACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUGGCC-AAGUGCU-- .(((((..(((---(((((-...))))))))(((----.......(((((....)))))(((((((....)))))))..........))))-))))...-- ( -27.30, z-score = -2.02, R) >droSim1.chrX 2935329 93 + 17042790 CACUUGCUGGCUGCUGGCG-UGGCGUCAGCUGCU----CGACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUGGGC-AAGUGCU-- (((((((((((.(((((((-...))))))).)))----.......(((((....)))))(((((((....)))))))...........)))-)))))..-- ( -35.20, z-score = -3.98, R) >droSec1.super_4 3411565 93 - 6179234 CACUUGCUGGCUGCUGGCG-UGGCGUCAGCUGCU----CGACUGGAGAUUUCUUAAUCUGUCGUUCUAAUGAAUGUCAUAAAUUUAUGGGC-AAGUGCU-- (((((((((((.(((((((-...))))))).)))----.(((.(((((((....))))).))((((....)))))))...........)))-)))))..-- ( -31.70, z-score = -2.76, R) >droYak2.chrX 17293771 85 - 21770863 CACUUGCCGGC---------UGGCGUCAGCUGCU----CGACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUGGGC-AAGUGCU-- (((((((((((---------((....))))).((----(.....)))...........((((((((....))))))))..........)))-)))))..-- ( -30.20, z-score = -3.57, R) >droEre2.scaffold_4690 1325337 85 + 18748788 CACUUGCUGGC---------UGGCGUCAGCUGCU----CGACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUGGGC-AAGUGCU-- (((((((((((---------....)))))).(((----((.....(((((....)))))(((((((....))))))).........)))))-)))))..-- ( -29.40, z-score = -3.55, R) >dp4.chrXL_group1e 12419357 94 + 12523060 UUUUUGGGGUCUGCUGCUACUGGCGUCAGCUGGA-----GACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUUGCCUAAGUGCU-- ..((((((((((.(..((.(..((....))..))-----)...).)))).........((((((((....))))))))...........))))))....-- ( -25.80, z-score = -1.77, R) >droPer1.super_14 1888298 94 + 2168203 UUUUUGGGGUCUGCUGCUACUGGCGUCAGCUGGA-----GACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUUGCCUAAGUGCU-- ..((((((((((.(..((.(..((....))..))-----)...).)))).........((((((((....))))))))...........))))))....-- ( -25.80, z-score = -1.77, R) >droWil1.scaffold_180588 137544 91 - 1294757 CACUUGCAGCA-------AUGGACGUCAGCUGCUUCUUCUUCAAGAGAUUUCUUAAUCUGACGCACUAAUGAAUGUCAUAAAUUUAUUGCC-AAGUGCC-- ((((((..(((-------(((((((((((........((((...)))).........)))))).....(((.....)))...)))))))))-)))))..-- ( -21.33, z-score = -1.28, R) >droGri2.scaffold_14853 2853696 83 - 10151454 -----GUUG----------CUGGCGUCAGCUGCUUAUUGGUUUAAGAAUUUCUUAAUCGGACAUUCUAAUGAAUGUCAUAAAUUGAUGGCGAUUUGGC--- -----...(----------(((....))))((((.((..(((((((.....))......(((((((....))))))).)))))..)))))).......--- ( -21.10, z-score = -1.79, R) >droVir3.scaffold_12928 6565665 75 + 7717345 ------CUGGC--------CUGGCGUCAGCUGCUUAU---GUUAAGGAUUUCUUAAUCGGGCAUUCUAAUGAAUGUCAUAAAUUGAUGGCGC--------- ------.....--------...((((((.....((((---((((((.....))))))..(((((((....))))))))))).....))))))--------- ( -18.90, z-score = -0.80, R) >droAna3.scaffold_13117 113084 76 - 5790199 --------------------UGGCGUCAGCUGCU----CGACUGGAGAUUUCCUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUGGCC-AAGUGCUGU --------------------......((((.(((----....((((((((....)))))(((((((....)))))))............))-)))))))). ( -19.10, z-score = -1.18, R) >consensus CACUUGCUGGC_________UGGCGUCAGCUGCU____CGACUGGAGAUUUCUUAAUCUGACGUUCUAAUGAAUGUCAUAAAUUUAUGGCC_AAGUGCU__ .....................(((....)))..............(((((....)))))(((((((....)))))))........................ (-11.08 = -11.49 + 0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:20 2011