
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,951,449 – 3,951,543 |
| Length | 94 |
| Max. P | 0.612582 |

| Location | 3,951,449 – 3,951,543 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 61.82 |
| Shannon entropy | 0.66810 |
| G+C content | 0.33426 |
| Mean single sequence MFE | -12.17 |
| Consensus MFE | -4.34 |
| Energy contribution | -4.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.558176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
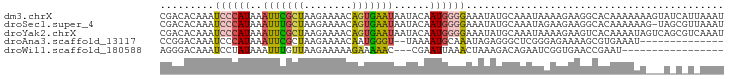
>dm3.chrX 3951449 94 + 22422827 CGACACAAAUCCCAUAAAUUCGCUAAGAAAACAGUGAAUAAUACAAUGGGGAAAUAUGCAAAUAAAAGAAGGCACAAAAAAAGUAUCAUUAAAU .((.((...((((((..(((((((........)))))))......)))))).....(((............)))........)).))....... ( -14.00, z-score = -2.63, R) >droSec1.super_4 3409261 93 - 6179234 CGACACAAAUCCCAUAAAUUUGCUAAGAAAACAGUGAAUAAUACAAUGGGGAAAUAUGCAAAUAGAAGAAGGCACAAAAAAG-UAGCGUUAAAU .(((.....((((((..(((..((........))..)))......))))))....................((((......)-).))))).... ( -12.00, z-score = -1.15, R) >droYak2.chrX 17290606 94 - 21770863 CGACACAAAUCCCAUAAAUUCGCUAAGAAAACAGUGAAUAAUACAAUGGGGAAAUAUGCAAAUAAAAGAAGUCACAAAAUAGUCAGCGUCAAAU .(((.(...((((((..(((((((........)))))))......))))))...(((....))).....................).))).... ( -14.40, z-score = -2.25, R) >droAna3.scaffold_13117 109915 78 - 5790199 CCGGACAAAUCCCAUAAAUUCGCUAAGAAAACAAUGGGU--UAAAAUGCAAAUAGAGGGCUCGGGAGAAAAGCGUGAAAU-------------- ((((......(((((...(((.....)))....))))).--......((.........))))))................-------------- ( -10.20, z-score = 0.93, R) >droWil1.scaffold_180588 135857 74 - 1294757 AGGGACAAAUCCUAUAAAUUUGUUAAGAAAAAGAAAAAC---CGAAUUAAACUAAAGACAGAAUCGGUGAACCGAAU----------------- .((((....)))).....((((((...............---..............)))))).((((....))))..----------------- ( -10.25, z-score = -1.30, R) >consensus CGACACAAAUCCCAUAAAUUCGCUAAGAAAACAGUGAAUAAUACAAUGGGGAAAUAUGCAAAUAAAAGAAGGCACAAAAAA___A_C_U_AAAU .........((((((..(((((((........)))))))......))))))........................................... ( -4.34 = -4.58 + 0.24)

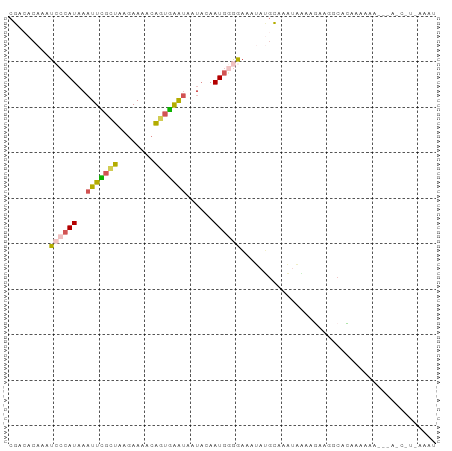
| Location | 3,951,449 – 3,951,543 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 61.82 |
| Shannon entropy | 0.66810 |
| G+C content | 0.33426 |
| Mean single sequence MFE | -13.60 |
| Consensus MFE | -4.22 |
| Energy contribution | -3.82 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
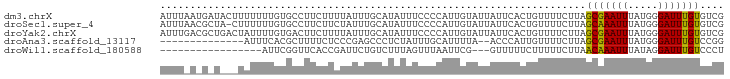
>dm3.chrX 3951449 94 - 22422827 AUUUAAUGAUACUUUUUUUGUGCCUUCUUUUAUUUGCAUAUUUCCCCAUUGUAUUAUUCACUGUUUUCUUAGCGAAUUUAUGGGAUUUGUGUCG .......(((((......(((((............)))))....(((((......((((.(((......))).))))..)))))....))))). ( -18.10, z-score = -3.64, R) >droSec1.super_4 3409261 93 + 6179234 AUUUAACGCUA-CUUUUUUGUGCCUUCUUCUAUUUGCAUAUUUCCCCAUUGUAUUAUUCACUGUUUUCUUAGCAAAUUUAUGGGAUUUGUGUCG .......((.(-(......))))............(((((..((((....(((..(((..(((......)))..))).)))))))..))))).. ( -12.10, z-score = -1.26, R) >droYak2.chrX 17290606 94 + 21770863 AUUUGACGCUGACUAUUUUGUGACUUCUUUUAUUUGCAUAUUUCCCCAUUGUAUUAUUCACUGUUUUCUUAGCGAAUUUAUGGGAUUUGUGUCG ....(((((.........((..(..........)..))......(((((......((((.(((......))).))))..)))))....))))). ( -17.30, z-score = -2.33, R) >droAna3.scaffold_13117 109915 78 + 5790199 --------------AUUUCACGCUUUUCUCCCGAGCCCUCUAUUUGCAUUUUA--ACCCAUUGUUUUCUUAGCGAAUUUAUGGGAUUUGUCCGG --------------.................(((((((...((((((.....(--((.....)))......))))))....))).))))..... ( -10.80, z-score = -0.11, R) >droWil1.scaffold_180588 135857 74 + 1294757 -----------------AUUCGGUUCACCGAUUCUGUCUUUAGUUUAAUUCG---GUUUUUCUUUUUCUUAACAAAUUUAUAGGAUUUGUCCCU -----------------....((...(((((..(((....)))......)))---))..............(((((((.....))))))).)). ( -9.70, z-score = -0.87, R) >consensus AUUU_A_G_U___UAUUUUGUGCCUUCUUCUAUUUGCAUAUUUCCCCAUUGUAUUAUUCACUGUUUUCUUAGCGAAUUUAUGGGAUUUGUGUCG .......................................................................(((((((.....))))))).... ( -4.22 = -3.82 + -0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:19 2011