| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,943,940 – 3,944,042 |
| Length | 102 |
| Max. P | 0.601570 |

| Location | 3,943,940 – 3,944,042 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.11 |
| Shannon entropy | 0.47288 |
| G+C content | 0.56269 |
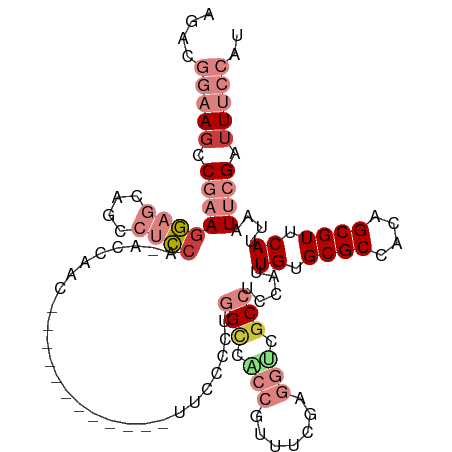
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -13.08 |
| Energy contribution | -15.94 |
| Covariance contribution | 2.86 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.601570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
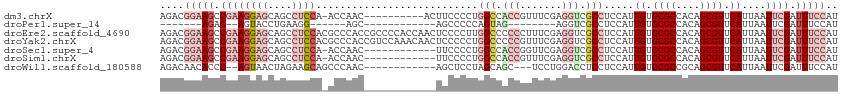
>dm3.chrX 3943940 102 + 22422827 AGACGGAAGCCGAAGGAGCAGCCUCCA-ACCAAC----------ACUUCCCCUGGCCACCGUUUCGAGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU ....((((..(((((((((.(((((.(-((....----------.((......)).....)))..))))).).))))..((.((((....)))).))....))))..)))).. ( -28.80, z-score = -1.80, R) >droPer1.super_14 1875658 78 + 2168203 -------AGAC--AGUACCUGAAGG------AGC------------AGCCCCCAGUAG--------AGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU -------....--......((((((------((.------------.((..((.....--------.))..))))))..((.((((....)))).))....))))........ ( -15.50, z-score = -0.35, R) >droEre2.scaffold_4690 1315561 113 + 18748788 AGACGGAAGCCGAAGGAGCAGCCUCCACGCCCACCGCCCCACCAACUCCCCUUGGCCCCCCUUUCGAGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU ....((((..(((((((((.(((((...((.....))....((((......))))..........))))).).))))..((.((((....)))).))....))))..)))).. ( -31.70, z-score = -2.35, R) >droYak2.chrX 17283251 113 - 21770863 AGACGGAAGCCGAAGGAGCAGCCUCCACGCCCACCGUCCAAACAACUCCCCCUGGCCCCCGUUUCGAGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU ....((((..(((((((((.(((((.(((....(((................)))....)))...))))).).))))..((.((((....)))).))....))))..)))).. ( -30.29, z-score = -1.54, R) >droSec1.super_4 3401859 100 - 6179234 AGACGGAAGCCGAAGGAGCAGCCUCCA-ACCAAC------------UUCCCCUGGCCACCGGUUCGAGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU ....((((..(((((((((.(((((.(-(((...------------..((...)).....)))).))))).).))))..((.((((....)))).))....))))..)))).. ( -33.80, z-score = -2.74, R) >droSim1.chrX 2926049 100 + 17042790 AGACGGAAGCCGAAGGAGCAGCCUCCA-ACCAAC------------UUCCCCUGGCCACCGUUUCGAGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU ....((((..(((((((((.(((((.(-((((..------------......))).......)).))))).).))))..((.((((....)))).))....))))..)))).. ( -28.41, z-score = -1.69, R) >droWil1.scaffold_180588 128042 96 - 1294757 AGACAACACCC--AGUAACUAGAAGCAGCCCAAC------------AGCUCCUAGCAGC---UCCUGGACCUCCUCCAUUGUGCGCCGCAGCGUUCAUUAAUUCGAUUUCCAU ...........--(((..((((.(((........------------.))).))))..))---)..((((..((......((.((((....)))).)).......))..)))). ( -17.22, z-score = -0.38, R) >consensus AGACGGAAGCCGAAGGAGCAGCCUCCA_ACCAAC____________UUCCCCUGGCCACCGUUUCGAGGUCGCCUCCAUUGUGCGCCACAGCGUUCAUUAAUUCGAUUUCCAU ....(((((.((((((((....))))...........................(((.(((.......))).))).....((.((((....)))).))....)))).))))).. (-13.08 = -15.94 + 2.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:16 2011