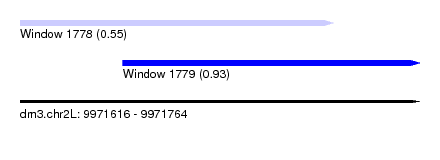
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,971,616 – 9,971,764 |
| Length | 148 |
| Max. P | 0.927593 |

| Location | 9,971,616 – 9,971,732 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 71.13 |
| Shannon entropy | 0.51707 |
| G+C content | 0.45016 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -14.65 |
| Energy contribution | -15.45 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
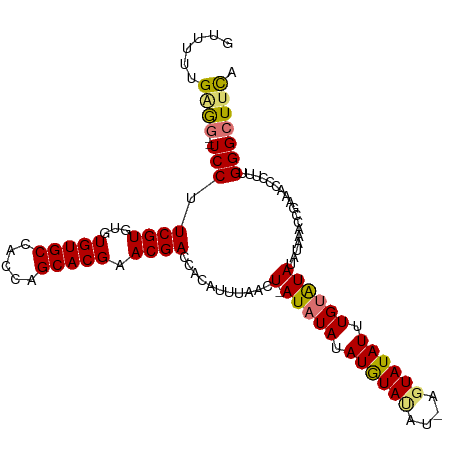
>dm3.chr2L 9971616 116 + 23011544 UGCUCCUGCGAGUUGAAUGUCCUUUCAGCUGGCCACGUGUUUUUGGGGCGUCCUUCGUGUGUGUGCCACCAGCACGAACGACCACAUUUAACU-AUCUAUAUGUAUAUAAGUAUAUU .(((((.((.(((((((......))))))).))...........)))))(((.((((((((((...)))..))))))).)))...........-......((((((....)))))). ( -31.00, z-score = -0.87, R) >droSec1.super_3 5418177 116 + 7220098 UCCCCCAGCGAGUUGAAUGUCCUUUCAGCUGGCCACGUGCUUUUGGAG-GUCCUUCGUGUGUGUGCCACCAGCACGAACGAACACAUUUAACUUAUAUAUAUAUAUUUUAGUAUAUU .........(((((((((((.(((((((..(((.....))).))))))-)...(((((...(((((.....))))).))))).)))))))))))....((((((......)))))). ( -29.60, z-score = -1.94, R) >droYak2.chr2L 12662326 113 + 22324452 UCCUUGGGCGAGUUGAAUGUCCUUUCAGCUGGCCACGUGUUUUUG-AG--UCCUUCGUGUGUGUGCCACCAGCACGAACGACCAGAUUUAACU-AUAUAUAUGUACAUGAGUAUAUU ......(((.(((((((......))))))).)))..((((..(((-((--((..((((...(((((.....))))).))))...)))))))..-))))..((((((....)))))). ( -35.30, z-score = -2.85, R) >droEre2.scaffold_4929 10591995 108 + 26641161 UCCUUGGGCGAGUUGAAUGUCCUUUUGGCUGGCCACGUGUUCUUG-AG-GUCCUUCGUGUGUGUGCCACCAGCACGAACGACCACAUUUAACU-AUAUAUAUGUAUAUAUU------ ......(((.(((..((......))..))).)))....(((..((-.(-(((.((((((((((...)))..))))))).)))).))...)))(-(((((....))))))..------ ( -32.40, z-score = -1.88, R) >droAna3.scaffold_12916 13810456 90 + 16180835 UCUUUCAACGAGUUGAAUGUCCUUUAAGC-GGCCUCGAGC---UGGGUCUCUUUUCGUGGGUAU---AUGGGUGUGUGUG----UGUAUGGCUGGAAUAUA---------------- ...(((((....)))))..(((....(((-(((((.....---.)))))......((((..(((---((......)))))----..)))))))))).....---------------- ( -17.30, z-score = 1.40, R) >consensus UCCUUCAGCGAGUUGAAUGUCCUUUCAGCUGGCCACGUGUUUUUGGAG_GUCCUUCGUGUGUGUGCCACCAGCACGAACGACCACAUUUAACU_AUAUAUAUGUAUAUAAGUAUAUU ..........((((((((((.......(((((.((((..(...(((((....))))).)..))))...)))))..........))))))))))........................ (-14.65 = -15.45 + 0.80)


| Location | 9,971,654 – 9,971,764 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 85.89 |
| Shannon entropy | 0.22955 |
| G+C content | 0.39739 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -18.05 |
| Energy contribution | -17.67 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.927593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9971654 110 + 23011544 GUUUUUGGGGCGUCCUUCGUGUGUGUGCCACCAGCACGAACGACCACAUUUAACU-AUCUAUAUGUAUAUAAGUAUAUUUGUGUAUAUAAAACGAAACCCUUUGGGCUUUA ((((..((((.(((.((((((((((...)))..))))))).)))...........-.....((((((((((((....))))))))))))........))))..)))).... ( -28.70, z-score = -1.60, R) >droSec1.super_3 5418215 109 + 7220098 GCUUUUGGAG-GUCCUUCGUGUGUGUGCCACCAGCACGAACGAACACAUUUAACUUAUAUAUAUAUAUUUUAGUAUAUUUGUGUAUAUAAC-CGAAACCCUUUGGGCUUCA .......(((-(((((((((...(((((.....))))).)))))..........(((((((((((.((........)).))))))))))).-...........))))))). ( -28.00, z-score = -2.36, R) >droYak2.chr2L 12662364 107 + 22324452 GUUUUUGAG---UCCUUCGUGUGUGUGCCACCAGCACGAACGACCAGAUUUAACU-AUAUAUAUGUACAUGAGUAUAUUUGUAUAUAGAACCCGAAACCCUUUGGGCUCCC ...((((.(---((.((((((((((...)))..))))))).))))))).....((-((((((((((((....))))))..))))))))..((((((....))))))..... ( -31.40, z-score = -4.24, R) >droEre2.scaffold_4929 10592033 102 + 26641161 GUUCUUGAGG--UCCUUCGUGUGUGUGCCACCAGCACGAACGACCACAUUUAACU-AUAUAUAUGUA------UAUAUUUGUAUAUGUAAACCGAAACCCUUUGGGCUUUC (((..((.((--((.((((((((((...)))..))))))).)))).))...))).-...((((((((------((....))))))))))....(((((((...))).)))) ( -28.40, z-score = -3.09, R) >consensus GUUUUUGAGG__UCCUUCGUGUGUGUGCCACCAGCACGAACGACCACAUUUAACU_AUAUAUAUGUAUAU_AGUAUAUUUGUAUAUAUAAACCGAAACCCUUUGGGCUUCA ((((..((((..((..((((...(((((.....))))).)))).............(((((.((((((....)))))).))))).........))...)))).)))).... (-18.05 = -17.67 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:23 2011