| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,931,056 – 3,931,170 |
| Length | 114 |
| Max. P | 0.963179 |

| Location | 3,931,056 – 3,931,170 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.02 |
| Shannon entropy | 0.35270 |
| G+C content | 0.33470 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.53 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
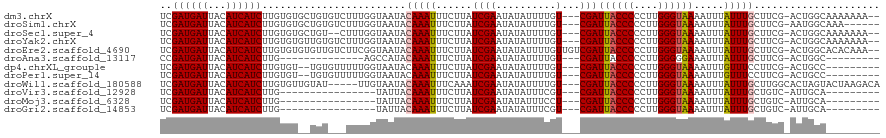
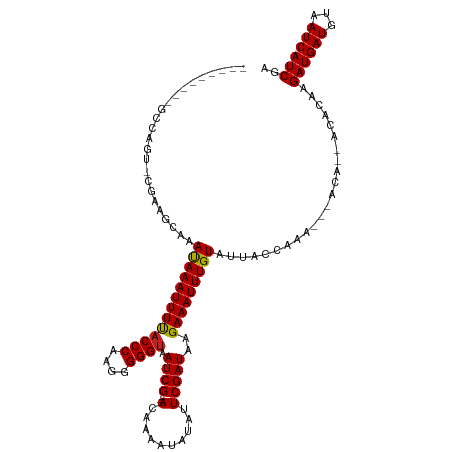
>dm3.chrX 3931056 114 + 22422827 UCGAUGAUUACAUCAUCUUGUGUGCUGUGUCUUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUCG-ACUGGCAAAAAAA-- ..((((((...)))))).....(((((.(((...(((((...((((((...(((((.((.....)))---))))(((((....)))))))))))..)))))..)-)))))))......-- ( -24.30, z-score = -2.20, R) >droSim1.chrX 2913003 110 + 17042790 UCGAUGAUUACAUCAUCUUGUGUGCUGUGUCUUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUCG-AAUGGCAAA------ ((((..(.(((((......)))))..((((.((((((((..........)))))))).)))).)..)---)))((((((....))))))......((((((...-...))))))------ ( -22.80, z-score = -1.58, R) >droSec1.super_4 3389318 112 - 6179234 UCGAUGAUUACAUCAUCUUGUGUGCUGU--CUUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUCG-ACUGGCAAAAAAA-- ..((((((...)))))).....((((((--(...(((((...((((((...(((((.((.....)))---))))(((((....)))))))))))..)))))..)-)).))))......-- ( -23.90, z-score = -2.31, R) >droYak2.chrX 17270799 114 - 21770863 UCGAUGAUUACAUCAUCUUGUGUGUUGUGUCUUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUCG-ACUGGCAAAAAAA-- ((((..(.(((((......)))))..((((.((((((((..........)))))))).)))).)..)---)))((((((....))))))......((((((...-...))))))....-- ( -23.70, z-score = -1.92, R) >droEre2.scaffold_4690 1301383 117 + 18748788 UCGAUGAUUACAUCAUCUUGUGUGUGUUGUCUUCGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGUUGUCGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUCG-ACUGGCACACAAA-- ..((((((...))))))((((((((..(((.((((((((..........)))))))).))).......(((((((((((....))))))............)))-))..)))))))).-- ( -31.20, z-score = -3.63, R) >droAna3.scaffold_13117 83723 93 - 5790199 CCGAUGAUUACAUCAUCUUG--------------AGCCAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGGGAAAUUUAUUUGCUUCG-ACUGGC--------- ((((((((...))))))(((--------------((.((...(((((((..(((((.((.....)))---))))..((((...)))))))))))...)).))))-)..)).--------- ( -21.20, z-score = -1.68, R) >dp4.chrXL_group1e 763741 105 - 12523060 UCGAUGAUUACAUCAUCUUGUGU--UGUGUUUUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUGUUUCCUUCG-ACUGCC--------- (((((((.((((......))))(--((((((......))))))).....))))))).........((---(((((((((....))))))............)))-))....--------- ( -22.90, z-score = -2.59, R) >droPer1.super_14 1859022 105 + 2168203 UCGAUGAUUACAUCAUCUUGUGU--UGUGUUUUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUGUUUCCUUCG-ACUGCC--------- (((((((.((((......))))(--((((((......))))))).....))))))).........((---(((((((((....))))))............)))-))....--------- ( -22.90, z-score = -2.59, R) >droWil1.scaffold_180588 114241 112 - 1294757 UCGAUGAUUACAUCAUCUUGUGUUGUAU-----UUGUAAUACAAAUUUCAAAUCGAAUAUAUUUUGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUGGCACUAGUACUAAGACA ..((((((...)))))).(((.((((((-----((((....(((((......((((.((.....)))---)))((((((....)))))).....)))))....)))..))))).)).))) ( -21.10, z-score = -1.10, R) >droVir3.scaffold_12928 6531786 91 + 7717345 UCGAUGAUUACAUCAUCUUG----------------UAUUACAAAUUUCUUAUCGAAUAUAUUUCGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUGUC-AUUGCA--------- (((((((.((((......))----------------))........(((.....)))......))))---)))((((((....))))))........(((....-...)))--------- ( -17.30, z-score = -2.18, R) >droMoj3.scaffold_6328 2201940 91 + 4453435 UCGAUGAUUACAUCAUCUUG----------------UAUUACAAAUUUCUUAUCGAAUAUAUUUCCU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUGUC-AUUGCA--------- .(((((((((((......))----------------))...(((((......((((..........)---)))((((((....)))))).....)))))..)))-))))..--------- ( -16.50, z-score = -2.38, R) >droGri2.scaffold_14853 2820317 91 - 10151454 UCGAUGAUUACAUCAUCUUG----------------UAUUACAAAUUUCUUAUCGAAUAUAUUUCGU---CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUGUC-AUUGCA--------- (((((((.((((......))----------------))........(((.....)))......))))---)))((((((....))))))........(((....-...)))--------- ( -17.30, z-score = -2.18, R) >consensus UCGAUGAUUACAUCAUCUUGUGU__UGU___UUUGGUAAUACAAAUUUCUUAUCGAAUAUAUUUUGU___CGAUUACCCCCUUGGGUAAAAUUUAUUUGCUUCG_ACUGGC_________ ..((((((...))))))........................(((((.......((((.....)))).......((((((....)))))).....)))))..................... (-11.60 = -11.53 + -0.07)
| Location | 3,931,056 – 3,931,170 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Shannon entropy | 0.35270 |
| G+C content | 0.33470 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.37 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.963179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
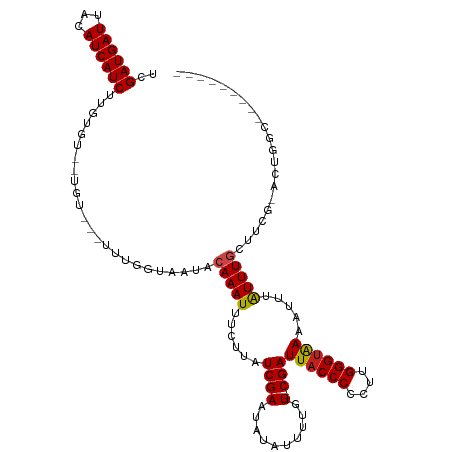
>dm3.chrX 3931056 114 - 22422827 --UUUUUUUGCCAGU-CGAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAAGACACAGCACACAAGAUGAUGUAAUCAUCGA --......(((..((-(.......(((((((((((((....)))).((((---(..........)))))..))))))))).........)))...))).....((((((...)))))).. ( -20.99, z-score = -1.71, R) >droSim1.chrX 2913003 110 - 17042790 ------UUUGCCAUU-CGAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAAGACACAGCACACAAGAUGAUGUAAUCAUCGA ------.((((....-....))))(((((((((((((....)))).((((---(..........)))))..))))))))).......................((((((...)))))).. ( -19.40, z-score = -1.48, R) >droSec1.super_4 3389318 112 + 6179234 --UUUUUUUGCCAGU-CGAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAAG--ACAGCACACAAGAUGAUGUAAUCAUCGA --......(((..((-(.......(((((((((((((....)))).((((---(..........)))))..))))))))).........)--)).))).....((((((...)))))).. ( -21.19, z-score = -1.75, R) >droYak2.chrX 17270799 114 + 21770863 --UUUUUUUGCCAGU-CGAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAAGACACAACACACAAGAUGAUGUAAUCAUCGA --..((((((...((-(((............((((((....)))))))))---))..(((((((((.....)))...))))))..))))))............((((((...)))))).. ( -20.80, z-score = -1.76, R) >droEre2.scaffold_4690 1301383 117 - 18748788 --UUUGUGUGCCAGU-CGAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCGACAACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCGAAGACAACACACACAAGAUGAUGUAAUCAUCGA --.(((((((...((-(((............((((((....)))))))))))...........((((.(((..........))).)))).......)))))))((((((...)))))).. ( -27.20, z-score = -2.80, R) >droAna3.scaffold_13117 83723 93 + 5790199 ---------GCCAGU-CGAAGCAAAUAAAUUUCCCCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUGGCU--------------CAAGAUGAUGUAAUCAUCGG ---------((((((-....))..((((((((((((....)))...((((---(..........)))))..))))))))).)))).--------------...((((((...)))))).. ( -24.20, z-score = -2.56, R) >dp4.chrXL_group1e 763741 105 + 12523060 ---------GGCAGU-CGAAGGAAACAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAAAACACA--ACACAAGAUGAUGUAAUCAUCGA ---------((..((-(((.(....).....((((((....)))))))))---))..(((((((((.....)))...)))))).)).........--......((((((...)))))).. ( -24.10, z-score = -3.44, R) >droPer1.super_14 1859022 105 - 2168203 ---------GGCAGU-CGAAGGAAACAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAAAACACA--ACACAAGAUGAUGUAAUCAUCGA ---------((..((-(((.(....).....((((((....)))))))))---))..(((((((((.....)))...)))))).)).........--......((((((...)))))).. ( -24.10, z-score = -3.44, R) >droWil1.scaffold_180588 114241 112 + 1294757 UGUCUUAGUACUAGUGCCAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACAAAAUAUAUUCGAUUUGAAAUUUGUAUUACAA-----AUACAACACAAGAUGAUGUAAUCAUCGA ((.(((.(((....))).))))).(((((((((((((....))))(((((---(..........)))))).))))))))).......-----...........((((((...)))))).. ( -20.50, z-score = -1.12, R) >droVir3.scaffold_12928 6531786 91 - 7717345 ---------UGCAAU-GACAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACGAAAUAUAUUCGAUAAGAAAUUUGUAAUA----------------CAAGAUGAUGUAAUCAUCGA ---------(((...-....))).(((((((((((((....)))).((((---(..........)))))..)))))))))....----------------...((((((...)))))).. ( -18.50, z-score = -2.10, R) >droMoj3.scaffold_6328 2201940 91 - 4453435 ---------UGCAAU-GACAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---AGGAAAUAUAUUCGAUAAGAAAUUUGUAAUA----------------CAAGAUGAUGUAAUCAUCGA ---------(((...-....))).(((((((((((((....)))).((((---((........))))))..)))))))))....----------------...((((((...)))))).. ( -20.70, z-score = -3.00, R) >droGri2.scaffold_14853 2820317 91 + 10151454 ---------UGCAAU-GACAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG---ACGAAAUAUAUUCGAUAAGAAAUUUGUAAUA----------------CAAGAUGAUGUAAUCAUCGA ---------(((...-....))).(((((((((((((....)))).((((---(..........)))))..)))))))))....----------------...((((((...)))))).. ( -18.50, z-score = -2.10, R) >consensus _________GCCAGU_CGAAGCAAAUAAAUUUUACCCAAGGGGGUAAUCG___ACAAAAUAUAUUCGAUAAGAAAUUUGUAUUACCAAA___ACA__ACACAAGAUGAUGUAAUCAUCGA ........................(((((((((((((....)))).((((...............))))..))))))))).......................((((((...)))))).. (-14.50 = -14.37 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:14 2011