
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,909,302 – 3,909,367 |
| Length | 65 |
| Max. P | 0.999072 |

| Location | 3,909,302 – 3,909,367 |
|---|---|
| Length | 65 |
| Sequences | 9 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.29640 |
| G+C content | 0.49528 |
| Mean single sequence MFE | -19.59 |
| Consensus MFE | -16.76 |
| Energy contribution | -16.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
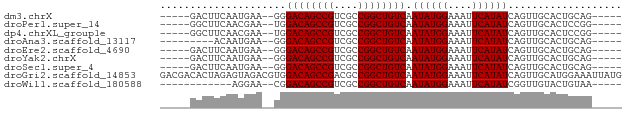
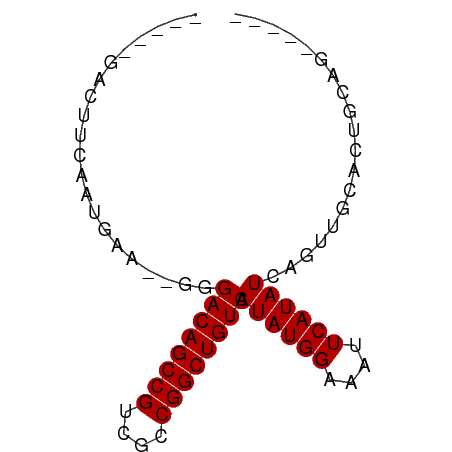
>dm3.chrX 3909302 65 + 22422827 -----GACUUCAAUGAA--GGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUGCAG----- -----..((((...)))--).((((((((....)))))))).((((((....))))))(((.....)))...----- ( -19.20, z-score = -1.79, R) >droPer1.super_14 1831436 65 + 2168203 -----GGCUUCAACGAA--UGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUCCGG----- -----((...((((...--..((((((((....)))))))).((((((....))))))..))))....))..----- ( -21.50, z-score = -2.43, R) >dp4.chrXL_group1e 737711 65 - 12523060 -----GGCUUCAACGAA--UGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUCCGG----- -----((...((((...--..((((((((....)))))))).((((((....))))))..))))....))..----- ( -21.50, z-score = -2.43, R) >droAna3.scaffold_13117 40960 61 - 5790199 ---------ACAAUGAA--GGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUGCAG----- ---------.((((...--..((((((((....)))))))).((((((....))))))..))))........----- ( -17.90, z-score = -1.70, R) >droEre2.scaffold_4690 1279878 65 + 18748788 -----GACUUCAAUGAA--GGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUGCAG----- -----..((((...)))--).((((((((....)))))))).((((((....))))))(((.....)))...----- ( -19.20, z-score = -1.79, R) >droYak2.chrX 17249252 65 - 21770863 -----GACUUCAAUGAA--GGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUGCAG----- -----..((((...)))--).((((((((....)))))))).((((((....))))))(((.....)))...----- ( -19.20, z-score = -1.79, R) >droSec1.super_4 3368157 65 - 6179234 -----GACUUCAAUGAA--GGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUGCAG----- -----..((((...)))--).((((((((....)))))))).((((((....))))))(((.....)))...----- ( -19.20, z-score = -1.79, R) >droGri2.scaffold_14853 2789851 77 - 10151454 GACGACACUAGAGUAGACGUGGACAGCCGACGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCAUGGAAAUUAUG .................((((((((((((....)))))))).((((((....))))))......))))......... ( -20.80, z-score = -1.79, R) >droWil1.scaffold_180588 91579 58 - 1294757 ------------AGGAA--CGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCGGUUGUACUGUAA----- ------------.(.((--(.((((((((....)))))))).((((((....))))))..))).).......----- ( -17.80, z-score = -1.74, R) >consensus _____GACUUCAAUGAA__GGGACAGCCGUCGCCGGCUGUCAAUAUGGAAAUUCAUAUCAGUUGCACUGCAG_____ .....................((((((((....)))))))).((((((....))))))................... (-16.76 = -16.76 + -0.00)
| Location | 3,909,302 – 3,909,367 |
|---|---|
| Length | 65 |
| Sequences | 9 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 84.58 |
| Shannon entropy | 0.29640 |
| G+C content | 0.49528 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
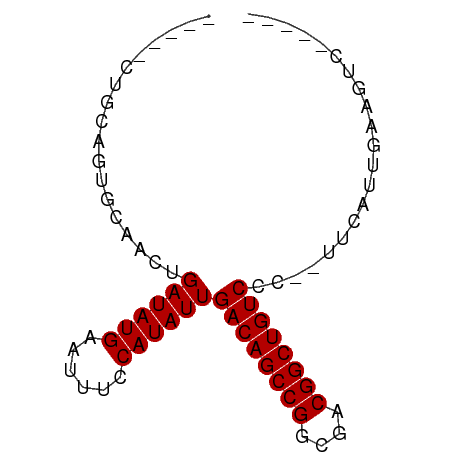
>dm3.chrX 3909302 65 - 22422827 -----CUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCC--UUCAUUGAAGUC----- -----((.(((((.....((((((......))))))((((((((....))))))))..--..))))).))..----- ( -24.10, z-score = -3.88, R) >droPer1.super_14 1831436 65 - 2168203 -----CCGGAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCA--UUCGUUGAAGCC----- -----..((....((((.((((((......))))))((((((((....))))))))..--...))))...))----- ( -25.60, z-score = -3.90, R) >dp4.chrXL_group1e 737711 65 + 12523060 -----CCGGAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCA--UUCGUUGAAGCC----- -----..((....((((.((((((......))))))((((((((....))))))))..--...))))...))----- ( -25.60, z-score = -3.90, R) >droAna3.scaffold_13117 40960 61 + 5790199 -----CUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCC--UUCAUUGU--------- -----..((((((.....((((((......))))))((((((((....))))))))..--..))))))--------- ( -24.70, z-score = -4.54, R) >droEre2.scaffold_4690 1279878 65 - 18748788 -----CUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCC--UUCAUUGAAGUC----- -----((.(((((.....((((((......))))))((((((((....))))))))..--..))))).))..----- ( -24.10, z-score = -3.88, R) >droYak2.chrX 17249252 65 + 21770863 -----CUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCC--UUCAUUGAAGUC----- -----((.(((((.....((((((......))))))((((((((....))))))))..--..))))).))..----- ( -24.10, z-score = -3.88, R) >droSec1.super_4 3368157 65 + 6179234 -----CUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCC--UUCAUUGAAGUC----- -----((.(((((.....((((((......))))))((((((((....))))))))..--..))))).))..----- ( -24.10, z-score = -3.88, R) >droGri2.scaffold_14853 2789851 77 + 10151454 CAUAAUUUCCAUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGUCGGCUGUCCACGUCUACUCUAGUGUCGUC ..................((((((......))))))((((((((....)))))))).(((.(.((....))).))). ( -19.50, z-score = -1.89, R) >droWil1.scaffold_180588 91579 58 + 1294757 -----UUACAGUACAACCGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCG--UUCCU------------ -----.............((((((......))))))((((((((....))))))))..--.....------------ ( -20.50, z-score = -4.36, R) >consensus _____CUGCAGUGCAACUGAUAUGAAUUUCCAUAUUGACAGCCGGCGACGGCUGUCCC__UUCAUUGAAGUC_____ ..................((((((......))))))((((((((....))))))))..................... (-20.11 = -20.11 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:10 2011