| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,897,257 – 3,897,349 |
| Length | 92 |
| Max. P | 0.788595 |

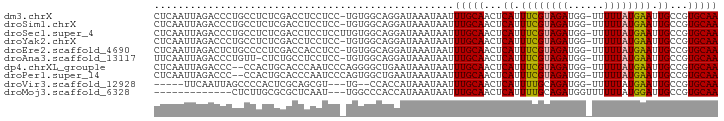
| Location | 3,897,257 – 3,897,349 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.63 |
| Shannon entropy | 0.40080 |
| G+C content | 0.42547 |
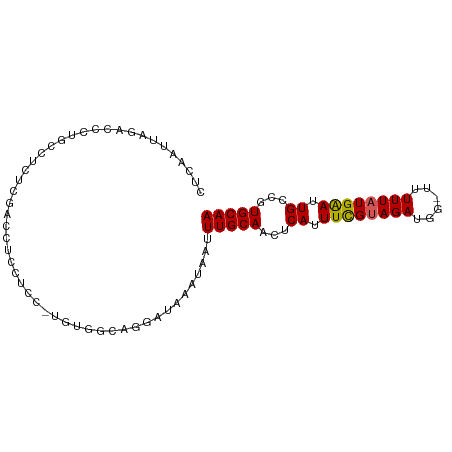
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -11.41 |
| Energy contribution | -11.36 |
| Covariance contribution | -0.05 |
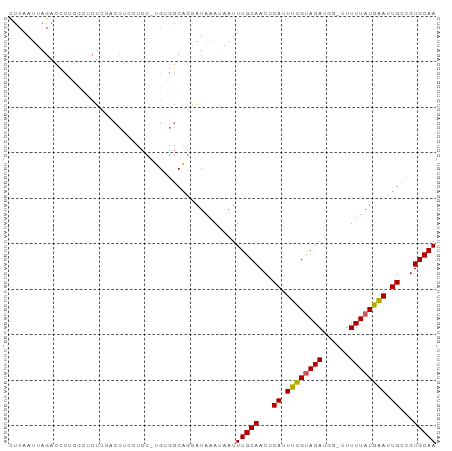
| Combinations/Pair | 1.13 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3897257 92 - 22422827 CUCAAUUAGACCCUGCCUCUCGACCUCCUCC-UGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA ...........((((((.(..((.....)).-.).)))))).........(((((...((.((((((((...-..)))))))).))...))))) ( -24.60, z-score = -2.79, R) >droSim1.chrX 2890102 92 - 17042790 CUCAAUUAGACCCUGCCUCUCGACCUCCUCC-UGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA ...........((((((.(..((.....)).-.).)))))).........(((((...((.((((((((...-..)))))))).))...))))) ( -24.60, z-score = -2.79, R) >droSec1.super_4 3356310 93 + 6179234 CUCAAUUAGACCCUGCCUCUCGACCUCCUCCUUGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA ...........((((((...(((........))).)))))).........(((((...((.((((((((...-..)))))))).))...))))) ( -23.90, z-score = -2.54, R) >droYak2.chrX 17237333 92 + 21770863 CUCAAUUAGACCCUGCCUCUCGACCUCCUCC-UGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA ...........((((((.(..((.....)).-.).)))))).........(((((...((.((((((((...-..)))))))).))...))))) ( -24.60, z-score = -2.79, R) >droEre2.scaffold_4690 1267690 92 - 18748788 CUCAAUUAGACUCUGCCCCUCGACCACCUCC-UGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA ...(((((.(..(((((.(..((.....)).-.).)))))..)...)))))((((...((.((((((((...-..)))))))).))...)))). ( -22.10, z-score = -1.81, R) >droAna3.scaffold_13117 24015 91 + 5790199 UUCAAUUAGACCCUGUU-CUCUGCCUCCUCC-UGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA ................(-(.(((((......-...)))))))........(((((...((.((((((((...-..)))))))).))...))))) ( -21.00, z-score = -1.63, R) >dp4.chrXL_group1e 723250 91 + 12523060 CUCAAUUAGACCC--CCACUGCACCCAAUCCCAGGGGCUGAAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA .....((((.(((--(.................)))))))).........(((((...((.((((((((...-..)))))))).))...))))) ( -20.73, z-score = -1.16, R) >droPer1.super_14 1816026 91 - 2168203 CUCAAUUAGACCC--CCACUGCACCCAAUCCCAGUGGCUGAAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG-UUUUUAUGAAUUGCCGUGCAA .....((((....--((((((..........)))))))))).........(((((...((.((((((((...-..)))))))).))...))))) ( -19.70, z-score = -1.78, R) >droVir3.scaffold_12928 6475261 83 - 7717345 -----UUCAAUUAGCCCCACUCGCAGCGU---UG--CCACCAUAAAUAAUUUGCAACUCAUUUUGCAGAUGG-UUUUUAUGAAUUGCCGUGCAA -----........((.......)).(((.---.(--(...((((((..((((((((......))))))))..-..))))))....))..))).. ( -17.00, z-score = -0.81, R) >droMoj3.scaffold_6328 2147217 78 - 4453435 -------------CUCUUGCGCGCUCAAU---UGGCCCACCAUAAAUAAUUUGCAACUCAUUUUGCAGAUGGUUUUUUAUGGAUUGCCGUGCAA -------------....(((((.......---.(((...(((((((..((((((((......)))))))).....)))))))...)))))))). ( -22.50, z-score = -2.10, R) >consensus CUCAAUUAGACCCUGCCUCUCGACCUCCUCC_UGUGGCAGGAUAAAUAAUUUGCAACUCAUUUCGUAGAUGG_UUUUUAUGAAUUGCCGUGCAA ..................................................(((((...((.((((((((......)))))))).))...))))) (-11.41 = -11.36 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:08 2011