| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,897,045 – 3,897,136 |
| Length | 91 |
| Max. P | 0.527971 |

| Location | 3,897,045 – 3,897,136 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.79 |
| Shannon entropy | 0.50730 |
| G+C content | 0.45679 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.86 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
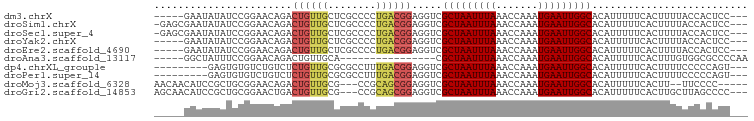
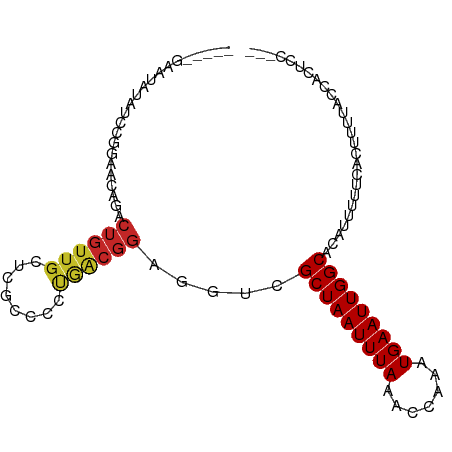
>dm3.chrX 3897045 91 - 22422827 -----GAAUAUAUCCGGAACAGACUGUUGCUCGCCCCUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUACCACUCC--- -----..........((....((((.(((.(((....)))))).))))(((((((((.......)))))))))................)).....--- ( -16.00, z-score = -0.51, R) >droSim1.chrX 2889885 95 - 17042790 -GAGCGAAUAUAUCCGGAACAGACUGUUGCUCGCCCCUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUACCACUCC--- -((((((.......(((......)))))))))(((((....)).))).(((((((((.......))))))))).......................--- ( -18.81, z-score = -0.72, R) >droSec1.super_4 3356093 95 + 6179234 -GAGCGAAUAUAUCCGGAACAGACUGUUGCUCGCCCCUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUACCACUCC--- -((((((.......(((......)))))))))(((((....)).))).(((((((((.......))))))))).......................--- ( -18.81, z-score = -0.72, R) >droYak2.chrX 17237121 91 + 21770863 -----GAAUAUAUCCGGAACAGACUGUUGCUCGCCCCUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUACCACUCC--- -----..........((....((((.(((.(((....)))))).))))(((((((((.......)))))))))................)).....--- ( -16.00, z-score = -0.51, R) >droEre2.scaffold_4690 1267478 91 - 18748788 -----GAAUAUAUCCGGAACAGACUGUUGCUCGCCCCUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUACCACUCC--- -----..........((....((((.(((.(((....)))))).))))(((((((((.......)))))))))................)).....--- ( -16.00, z-score = -0.51, R) >droAna3.scaffold_13117 23780 78 + 5790199 -----GGCUAUUUCCGGAACAGACUGUUGCA----------------CGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUGUGGCGCCCCAA -----.(((((....(.(((.....))).).----------------.(((((((((.......)))))))))..............)))))....... ( -15.20, z-score = -0.10, R) >dp4.chrXL_group1e 722991 87 + 12523060 ---------GAGUGUGUCUGUCUCUGUUGCGCGCCUUUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUCCCCCAGU--- ---------(((((....(((((((((((........))))))))...(((((((((.......)))))))))))).....)))))..........--- ( -19.30, z-score = -0.44, R) >droPer1.super_14 1815761 87 - 2168203 ---------GAGUGUGUCUGUCUCUGUUGCGCGCCUUUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUCCCCCAGU--- ---------(((((....(((((((((((........))))))))...(((((((((.......)))))))))))).....)))))..........--- ( -19.30, z-score = -0.44, R) >droMoj3.scaffold_6328 2146982 89 - 4453435 AACAACAUCCGCUGCGGAACAGACUGUUGCG---CCGCAGCGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUU--UUCCCC----- ....((.((((((((((..(((....)))..---)))))))))).)).(((((((((.......))))))))).............--......----- ( -31.40, z-score = -4.30, R) >droGri2.scaffold_14853 2774009 93 + 10151454 AGCAACAUCCGCUGCGGAACUGACUGUUGCG---CCGCAGCGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUGCUUAGCCCC--- (((((..((((((((((..(........)..---)))))))))).((.(((((((((.......))))))))))).........))))).......--- ( -33.80, z-score = -3.49, R) >consensus _____GAAUAUAUCCGGAACAGACUGUUGCUCGCCCCUGACGGAGGUCGCUAAUUUAAACCAAAUGAAUUGGCACAUUUUUCACUUUUACCACUCC___ .......................((((((........)))))).....(((((((((.......))))))))).......................... (-10.68 = -10.86 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:07 2011