
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,888,182 – 3,888,238 |
| Length | 56 |
| Max. P | 0.990622 |

| Location | 3,888,182 – 3,888,238 |
|---|---|
| Length | 56 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | forward |
| Mean pairwise identity | 62.68 |
| Shannon entropy | 0.70414 |
| G+C content | 0.61764 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -12.27 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
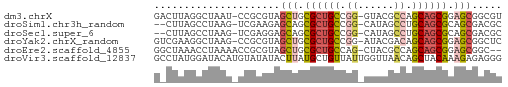
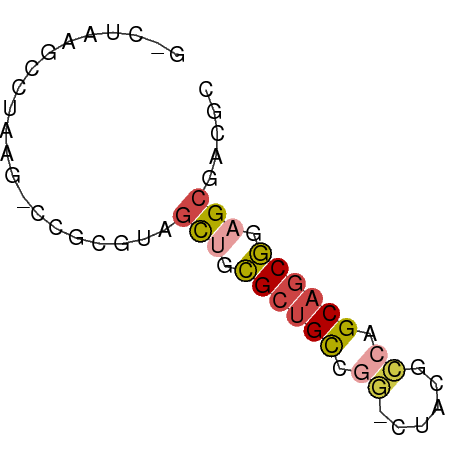
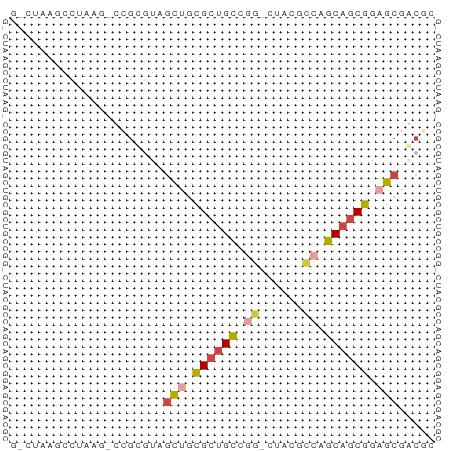
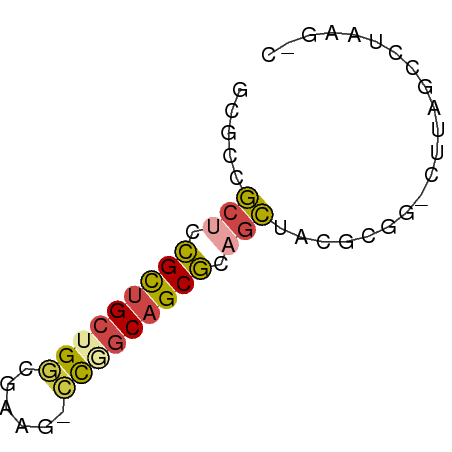
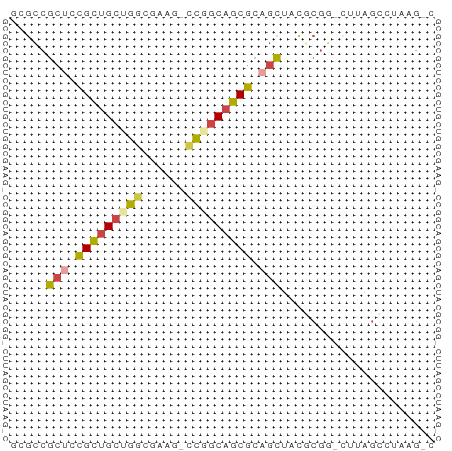
>dm3.chrX 3888182 56 + 22422827 GACUUAGGCUAAU-CCGCGUAGCUGCGCUGCCGG-GUACGCCAGCAGCGGAGCGGCGU ......((.....-))((((.(((.((((((.((-.....)).)))))).))).)))) ( -24.80, z-score = -0.54, R) >droSim1.chr3h_random 107942 54 - 1452968 --CUUAGCCUAAG-UCGAAGAGCAGCGCUGCCGG-CAUAGCCUGCAGCGCAGCGACGC --..........(-(((.......(((((((.((-(...))).)))))))..)))).. ( -22.50, z-score = -1.28, R) >droSec1.super_6 3669715 54 + 4358794 --CUUAGCCUAAG-UCGAGGAGCAGCGCUGCCGG-CAUAGCCUGCAGCGCAGCGACGC --.....(((...-...))).((.(((((((.((-(...))).))))))).))..... ( -23.50, z-score = -1.19, R) >droYak2.chrX_random 539170 56 + 1802292 GUCGAAGGCUAAG-CCGCGUAGCUGCGCUGCCGG-AUACGACAGCAGCGGAGCGGCUC ...........((-((((....((((((((.((.-...)).)))).)))).)))))). ( -25.30, z-score = -1.23, R) >droEre2.scaffold_4855 7559 55 + 242605 GGCUAAACCUAAAACCGCGUAGCUGCGCUGCCAG-CUACGCCAGCAGCGGAGCGGC-- ..............(((((((((((......)))-))))((.....))...)))).-- ( -18.60, z-score = 0.55, R) >droVir3.scaffold_12837 5114 58 - 117923 GCCUAUGGAUACAUGUAUAUACUUAUGCUGUUAUUGGUUAACAGCUACAAAGAGAGGG .((((((....))).......(((..(((((((.....)))))))....)))..))). ( -12.40, z-score = -0.91, R) >consensus G_CUAAGCCUAAG_CCGCGUAGCUGCGCUGCCGG_CUACGCCAGCAGCGGAGCGACGC .....................(((.((((((.((......)).)))))).)))..... (-12.27 = -12.83 + 0.56)
| Location | 3,888,182 – 3,888,238 |
|---|---|
| Length | 56 |
| Sequences | 6 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 62.68 |
| Shannon entropy | 0.70414 |
| G+C content | 0.61764 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.990622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
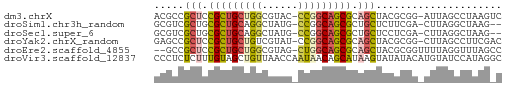
>dm3.chrX 3888182 56 - 22422827 ACGCCGCUCCGCUGCUGGCGUAC-CCGGCAGCGCAGCUACGCGG-AUUAGCCUAAGUC .(((.(((.(((((((((.....-))))))))).)))...))).-............. ( -24.60, z-score = -1.39, R) >droSim1.chr3h_random 107942 54 + 1452968 GCGUCGCUGCGCUGCAGGCUAUG-CCGGCAGCGCUGCUCUUCGA-CUUAGGCUAAG-- ..(((((.(((((((.(((...)-)).))))))).)).....))-)..........-- ( -22.50, z-score = -0.50, R) >droSec1.super_6 3669715 54 - 4358794 GCGUCGCUGCGCUGCAGGCUAUG-CCGGCAGCGCUGCUCCUCGA-CUUAGGCUAAG-- ..(((((.(((((((.(((...)-)).))))))).)).....))-)..........-- ( -22.50, z-score = -0.35, R) >droYak2.chrX_random 539170 56 - 1802292 GAGCCGCUCCGCUGCUGUCGUAU-CCGGCAGCGCAGCUACGCGG-CUUAGCCUUCGAC (((((((...((((((((((...-.)))))..)))))...))))-))).......... ( -25.70, z-score = -2.09, R) >droEre2.scaffold_4855 7559 55 - 242605 --GCCGCUCCGCUGCUGGCGUAG-CUGGCAGCGCAGCUACGCGGUUUUAGGUUUAGCC --((.(((.((((((..((...)-)..)))))).)))...))((((........)))) ( -24.00, z-score = -0.55, R) >droVir3.scaffold_12837 5114 58 + 117923 CCCUCUCUUUGUAGCUGUUAACCAAUAACAGCAUAAGUAUAUACAUGUAUCCAUAGGC .(((....((((.(((((((.....)))))))))))(((((....)))))....))). ( -9.80, z-score = -1.06, R) >consensus GCGCCGCUCCGCUGCUGGCGAAG_CCGGCAGCGCAGCUACGCGG_CUUAGCCUAAG_C .....(((.(((((((((......))))))))).)))..................... (-14.80 = -14.92 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:06 2011