
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,870,026 – 3,870,119 |
| Length | 93 |
| Max. P | 0.539580 |

| Location | 3,870,026 – 3,870,119 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.22 |
| Shannon entropy | 0.57345 |
| G+C content | 0.54842 |
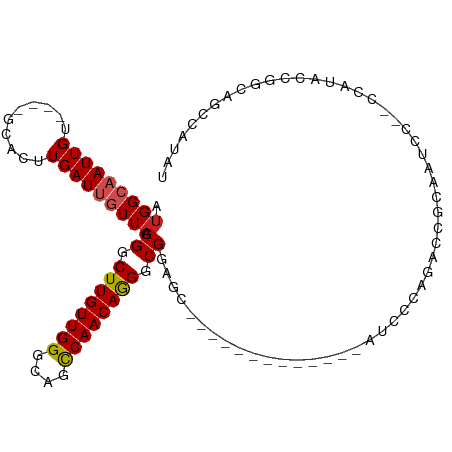
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -16.85 |
| Energy contribution | -17.07 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.539580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
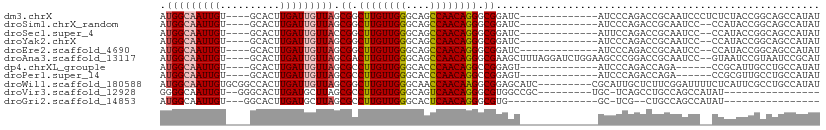
>dm3.chrX 3870026 93 - 22422827 AUGGCAAUUGU----GCACUUGAUUGUUAGCGGCUUGUUGGGCAGCCAACAGGGCGGAUC-------------AUCCCAGACCGCAAUCCCUCUCUACCGGCAGCCAUAU (((((.....(----((((......))...(((((((((((....))))))))((((.((-------------......))))))............))))))))))).. ( -28.70, z-score = 0.14, R) >droSim1.chrX_random 1520932 91 - 5698898 AUGGCAAUUGU----GCACUUGAUUGUUAGCGGCUUGUUGGGCAGCCAACAGGGCGGAUC-------------AUCCCAGACCGCAAUCC--CCAUACCGGCAGCCAUAU (((((.....(----((((......))...(((((((((((....))))))))((((.((-------------......)))))).....--.....))))))))))).. ( -28.70, z-score = 0.11, R) >droSec1.super_4 3331373 91 + 6179234 AUGGCAAUUGU----GCACUUGAUUGUUACCGGCUUGUUGGGCAGCCAACAGGGCGGAUC-------------AUUCCAGACCGCAAUCC--CCAUACCGGCAGCCAUAU (((((.(((((----(..((.((.((...(((.((((((((....)))))))).)))..)-------------).)).))..))))))..--((.....))..))))).. ( -33.00, z-score = -1.63, R) >droYak2.chrX 17217638 91 + 21770863 AUGGCAAUUGU----GCACUUGAUUGUUAGCGGCUUGUUGGGCAGCCAACAGGGCGGAUC-------------AUCCCAGACCGCAAUCC--CCAUACCGGCAGCCAUAU (((((.....(----((((......))...(((((((((((....))))))))((((.((-------------......)))))).....--.....))))))))))).. ( -28.70, z-score = 0.11, R) >droEre2.scaffold_4690 1245151 91 - 18748788 AUGGCAAUUGU----GCACUUGAUUGUUAGCGGCUUGUUGGGCAGCCAACAGGGCGGAUC-------------AUCCCAGACCGCAAUCC--CCAUACCGGCAGCCAUAU (((((.....(----((((......))...(((((((((((....))))))))((((.((-------------......)))))).....--.....))))))))))).. ( -28.70, z-score = 0.11, R) >droAna3.scaffold_13117 1084 104 + 5790199 AUGGCAAUUGU----GCACUUGAUUGUUAGCGACUUGUUGGGCAGCCAACAGGGCGAAGCUUUAGGAUCUGGAAGCCCGGACCGCAAUCC--GUAAUCCGUAAUCCGCAU .(((((((((.----.....)))))))))(((.((((((((....))))))))(((........((.(((((....)))))))((.....--))....)))....))).. ( -30.70, z-score = 0.14, R) >dp4.chrXL_group1e 698412 87 + 12523060 AUGGCAAUUGU----GCACUUGAUUGUUAGCGCCUUGUUGGGCACCCAACAGGCCGGAGU-------------AUCCCAGACCAGA------CCGCAUUGCCUGCCAUAU ((((((...((----((............(.((((.(((((....))))))))))((...-------------...))........------..))))....)))))).. ( -28.80, z-score = -1.25, R) >droPer1.super_14 1790627 87 - 2168203 AUGGCAAUUGU----GCACUUGAUUGUUAGCGCCUUGUUGGGCACCCAACAGGCCGGAGU-------------AUCCCAGACCAGA------CCGCGUUGCCUGCCAUAU ((((((.....----(((......)))((((((((((((((....))))))))..((...-------------...))........------..))))))..)))))).. ( -26.50, z-score = -0.32, R) >droWil1.scaffold_180588 57442 101 + 1294757 AUGGCAAUUGUGCGGCCACUUGAUUGUUAGCGGCUUGUUGGGCAACCAACAAGGCGGAGCAUC---------CGCAUUGCUCUUCGGAUUUUCUCAUUCGCCUGCCAUAU (((((........((((.((........)).))))((((((....))))))((((((((((..---------.....))))))..((((......))))))))))))).. ( -38.10, z-score = -2.29, R) >droVir3.scaffold_12928 6447465 82 - 7717345 GGGGCAAUUGU--GGGCACUUGAUGCUUAGCGCCUUGUUGGGCAGUCAACAGGGCGUGGCCGC---------UGC-UCAGCCUGCCAGCCAUAU---------------- ..(((.....(--(((((.....))))))((((((((((((....))))))))))))(((.((---------(..-..)))..))).)))....---------------- ( -35.00, z-score = -0.40, R) >droGri2.scaffold_14853 2740666 73 + 10151454 AUGGCAAUUGU---GGCACUUGAUGCUUAGCGCCUUGUUGGGCACUCAACAGGGCGUG---------------GC-UCG--CUGCCAGCCAUAU---------------- (((((.....(---((((..(((.(((..((((((((((((....)))))))))))))---------------))-)))--.))))))))))..---------------- ( -35.20, z-score = -3.16, R) >consensus AUGGCAAUUGU____GCACUUGAUUGUUAGCGGCUUGUUGGGCAGCCAACAGGGCGGAGC_____________AUCCCAGACCGCAAUCC__CCAUACCGGCAGCCAUAU .(((((((((..........))))))))).((.((((((((....)))))))).))...................................................... (-16.85 = -17.07 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:02 2011