| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,858,256 – 3,858,356 |
| Length | 100 |
| Max. P | 0.596982 |

| Location | 3,858,256 – 3,858,356 |
|---|---|
| Length | 100 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.18 |
| Shannon entropy | 0.68847 |
| G+C content | 0.54182 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -9.97 |
| Energy contribution | -9.66 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
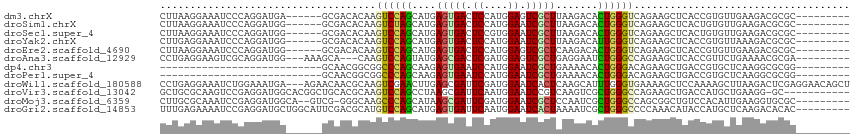
>dm3.chrX 3858256 100 - 22422827 CUUAAGGAAAUCCCAGGAUGA------GCGACACAAGUCCAGCAUGAGUGACUCCAUGGAGUCGCUUAAGACACUGGGUCAGAAGCUCACCGUGUUGAAGACGCGC--------- .....((.....)).((.(((------(((((....))).....((((((((((....)))))))))).(((.....)))....)))))))(((((...)))))..--------- ( -34.00, z-score = -2.09, R) >droSim1.chrX 2872096 100 - 17042790 CUUAAGGAAAUCCCAGGAUGG------GCGACACAAGUCUAGCAUGAGUGACUCCAUGGAAUCGCUUAAGACACUGGGUCAGAAGCUCACUGUGUUGAAGACGCGC--------- (((........(((.....))------)(((((((.((((....(((((((.((....)).)))))))))))..(((((.....))))).))))))))))......--------- ( -27.40, z-score = -0.23, R) >droSec1.super_4 3319798 100 + 6179234 CUUAAGGAAAUCCCAGGAUGG------GCGACACAAGUCCAGCAUGAGUGACUCCGUGGAAUCGCUUAAGACACUGGGUCAGAAGCUCACUGUGUUGAAGACGCGC--------- (((...((...(((((..(((------((.......)))))...(((((((.((....)).))))))).....)))))))..)))......(((((...)))))..--------- ( -27.80, z-score = 0.11, R) >droYak2.chrX 17205752 100 + 21770863 CUUGAGGAAAUCCCAGGAUGG------GCGACACAAGUCCAGCAUGAGUGACUCCAUGGAAUCGCUUAAGACAUUGGGUCAGAAGCUCACCGUGUUAAAGACGCGC--------- ((((.((....)))))).(((------((.......)))))((.(((((((.((....)).))))))).((((((((((.....)))))..)))))......))..--------- ( -30.00, z-score = -1.04, R) >droEre2.scaffold_4690 1233173 100 - 18748788 CUUAAGGAAAUCCCAGGAUGG------GCGACACAAGUCCAGCAUGAGUGACUCCAUGGAGUCGCUCAAGACACUGGGUCAGAAGCUCACCGUGUUGAAGACGCGC--------- (((...((...(((((..(((------((.......)))))...((((((((((....)))))))))).....)))))))..))).....((((((...)))))).--------- ( -37.40, z-score = -2.40, R) >droAna3.scaffold_12929 2972277 100 - 3277472 CCUGAGGAAGUCGCAGGAUGG---AAAGCA---CAAGUCCAGUAUGAGCGACUCGAUGGAGUCGCUGAGGAAUCUGGGCCAGAAGCUCACCGUUCUGAAAACGCGA--------- .........((..((((((((---..(((.---...((((((..(.((((((((....)))))))).).....)))))).....)))..))))))))...))....--------- ( -38.50, z-score = -2.49, R) >dp4.chr3 9771257 79 - 19779522 ---------------------------GCAACGGCGGCCCAGCAAGAGUGAAUCCAUGGAAUCGCUGAAAACACUGGGACAGAAGCUGACCGUGCUCAAGGCGCGG--------- ---------------------------....((((.((((((....(((((.((....)).))))).......))))).)....)))).((((((.....))))))--------- ( -29.10, z-score = -1.90, R) >droPer1.super_4 5091814 79 - 7162766 ---------------------------GCAACGGCGGCCCAGCAAGAGUGAAUCCAUGGAAUCGCUGAAAACACUGGGACAGAAGCUGACCGUGCUCAAGGCGCGG--------- ---------------------------....((((.((((((....(((((.((....)).))))).......))))).)....)))).((((((.....))))))--------- ( -29.10, z-score = -1.90, R) >droWil1.scaffold_180588 48204 112 + 1294757 CCUGAGGAAAUCUGGAAAUGA---AGAACAACGCAAGUCGAACUUGAGCGAUUCGAUGGAAUCACUCAAGCAUUUGGGUGAAAAGCUCCAAAAGCUUAAGACUCGAGGAACAGCU ((((((...(((..((.....---....(.((....)).)..((((((.(((((....))))).))))))..))..)))...(((((.....)))))....))).)))....... ( -30.30, z-score = -1.65, R) >droVir3.scaffold_13042 3041490 103 + 5191987 GCUGCGCAAGUCCGAGGAUGGCACGGCUGCACGCAAGUCCAGCCUAAGCGAUUCAAUGGAAUCCGUCAAGUCGCUGGGCCAGAAGCUGACCAUGCUGAAGG-GC----------- (((.((((.(((.(....((((..(((((.((....)).)))))..(((((((..((((...))))..)))))))..))))....).)))..))).)...)-))----------- ( -38.10, z-score = -0.47, R) >droMoj3.scaffold_6359 2472052 103 + 4525533 CUUGCGCAAAUCCGAGGAUGGCA--GUCG-GGGCAAGCCCAGCAUAAGCGAUUCGAUGGAAUCGCUCCAAUCGCUGGGCCAGCGGCUGUCCACAUUGAAGGUGCGC--------- ...(((((..(.(((...(((((--((((-..(...(((((((...((((((((....))))))))......))))))))..)))))).)))..))).)..)))))--------- ( -46.10, z-score = -2.63, R) >droGri2.scaffold_14853 6707116 106 + 10151454 UUUGAGAAAAUCCGAGGAUGCUGGCAUUCGACGCAUGUCCAGCAUGAGUGAUUCAAUGGAAUCACUAAAAUCGCUGGGCCCCAAACAUACCAUGCUCAAGACACAC--------- ((((((.........(((((....))))).......(((((((((.((((((((....))))))))...)).)))))))...............))))))......--------- ( -32.00, z-score = -2.56, R) >consensus CUUGAGGAAAUCCCAGGAUGG______GCGACGCAAGUCCAGCAUGAGUGACUCCAUGGAAUCGCUCAAGACACUGGGUCAGAAGCUCACCGUGCUGAAGACGCGC_________ ....................................((((((....(((((.((....)).))))).......)))))).................................... ( -9.97 = -9.66 + -0.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:00 2011