
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,845,686 – 3,845,786 |
| Length | 100 |
| Max. P | 0.949858 |

| Location | 3,845,686 – 3,845,786 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Shannon entropy | 0.50844 |
| G+C content | 0.45380 |
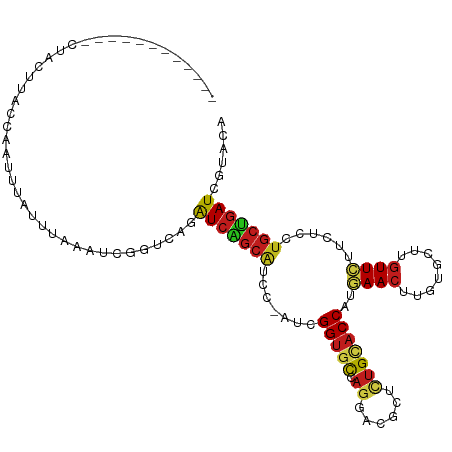
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -14.98 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
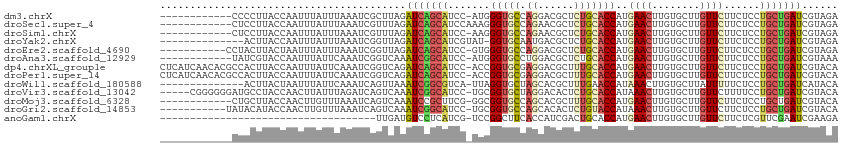
>dm3.chrX 3845686 100 - 22422827 ------------CCCCUUACCAAUUUAUUUAAAUCGCUUAGAUCAGCAUCC-AUGGGUGCCAGGACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUAGA ------------............................((((((((...-...(((((.(((....))))))))..((((........))))......))))))))..... ( -25.70, z-score = -1.86, R) >droSec1.super_4 3307220 101 + 6179234 ------------CUCCUUACCAAUUUAUUUAAAUCGUUUAGAUCAGCAUCCAAAGGGUGCCAGAACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUAGA ------------............................((((((((.......(((((.(((....))))))))..((((........))))......))))))))..... ( -26.60, z-score = -2.68, R) >droSim1.chrX 2859229 100 - 17042790 ------------CUCCUUACCAAUUUAUUUAAAUCGUUUAGAUCAGCAUCC-AAGGGUGCCAGAACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUAGA ------------............................((((((((...-...(((((.(((....))))))))..((((........))))......))))))))..... ( -26.60, z-score = -2.66, R) >droYak2.chrX 17193778 98 + 21770863 --------------ACUUACCAAUUUAUUUAAAUCGGUUAGAUCAGCAUCGUAU-GGUGCAAUGACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUAGA --------------....(((.((((....)))).)))..((((((((....((-((((((.........))))))))((((........))))......))))))))..... ( -29.00, z-score = -3.30, R) >droEre2.scaffold_4690 1221206 101 - 18748788 -----------CCUACUUACUAAUUUAUUUAAAUCGGUUAGAUCAGCAUCC-GUGGGUGCCAGGACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUAGA -----------.((((..((..((((....))))..))..((((((((...-...(((((.(((....))))))))..((((........))))......)))))))))))). ( -30.50, z-score = -2.89, R) >droAna3.scaffold_12929 2959594 100 - 3277472 ------------UAUCGUACCAAUUUAUUCAAAUCGGUCAAAUCGGCAUCC-AUGGGUGCCUGGACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUAAA ------------......................((((((....((((((.-...)))))).(((((....((((.........)))).))))).........)))))).... ( -23.20, z-score = -1.00, R) >dp4.chrXL_group1e 681776 112 + 12523060 CUCAUCAACACGCCACUUACCAAUUUAUUCAAAUCGGUCAGAUCAGCAUCC-ACCGGUGCGAGGACGCUUUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUACA .........((.......(((.((((....)))).)))..((((((((...-...(((((((((...)))))))))..((((........))))......))))))))))... ( -30.80, z-score = -2.80, R) >droPer1.super_14 1773415 112 - 2168203 CUCAUCAACACGCCACUUACCAAUUUAUUCAAAUCGGUCAGAUCAGCAUCC-ACCGGUGCGAGGACGCUUUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUACA .........((.......(((.((((....)))).)))..((((((((...-...(((((((((...)))))))))..((((........))))......))))))))))... ( -30.80, z-score = -2.80, R) >droWil1.scaffold_180588 37055 98 + 1294757 --------------ACUUACUAAUUUAUUCAAAUCAGUUAAAUCGGCGUCA-UUAGGUGCUAGCACGCUUUGAACCAUAAACUUGUGCUUAUUUUUCUCCUGCUGAUCAUACA --------------..................((((((......(((..(.-...)..)))((((((..((........))..))))))............))))))...... ( -15.10, z-score = -0.38, R) >droVir3.scaffold_13042 4752682 107 - 5191987 -----CGGGGGGAUGCCUACCAACUUAUUUAGAUCAGUCAAAUCGGCAUCC-UGCGGUGCUAGGACACUCUGCACCAUAAACUUGUGCUUGUUCUUUUCCUGCUGAUCGUACA -----.((..((...))..))..........(((((((......((((((.-...))))))((((((....((((.........)))).))))))......)))))))..... ( -28.30, z-score = -0.62, R) >droMoj3.scaffold_6328 3999599 100 - 4453435 ------------CUGCUUACCAACUUGUUUAAAUCAGUCAAAUCCGCUUCG-GGCGGUGCCAGCACGCUUUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUACA ------------.............(((....((((((....((((...))-)).(((((.((....))..)))))..((((........)))).......))))))...))) ( -19.40, z-score = 0.79, R) >droGri2.scaffold_14853 9819627 101 + 10151454 -----------UAUACAUACCAACUUGUUUAAAUCAGUCAAAUCGGCAUCC-UGCGGUGCCAGCACACUCUGUACCAUAAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUACA -----------..............(((....((((((......((((((.-...))))))((((((....((.......)).))))))............))))))...))) ( -20.30, z-score = -1.30, R) >anoGam1.chrX 10868765 76 + 22145176 ------------------------------------UUGAUGUCCUCAUCG-UCCGGCUUCACCAUCGACUGCACCAUGAACUUGUGCUUGUUCUUCUCGUUCGAAUCGAAGA ------------------------------------..((((....)))).-.....((((....((((.((......((((........))))....)).))))...)))). ( -12.50, z-score = 0.95, R) >consensus ____________CUACUUACCAAUUUAUUUAAAUCGGUCAGAUCAGCAUCC_AUCGGUGCCAGGACGCUCUGCACCAUGAACUUGUGCUUGUUCUUCUCCUGCUGAUCGUACA .........................................(((((((.......(((((.((......)))))))..((((........))))......)))))))...... (-14.98 = -15.22 + 0.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:13:00 2011