
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,808,494 – 3,808,587 |
| Length | 93 |
| Max. P | 0.822341 |

| Location | 3,808,494 – 3,808,587 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.43041 |
| G+C content | 0.44569 |
| Mean single sequence MFE | -23.46 |
| Consensus MFE | -13.36 |
| Energy contribution | -14.48 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.564180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3808494 93 + 22422827 AAUUUCGGUAGCAAAC-GCACUUCCCUUGUCUCGCCUAUCCAUCAUUUUGGUUAGGCGCACAGUCGCGGUUACCAUAUUAGUGAAAUCUUGUUA-------------------------- ......((((((...(-(((((......((..((((((.(((......))).)))))).))))).)))))))))....................-------------------------- ( -24.00, z-score = -2.50, R) >droSim1.chrX 2803973 91 + 17042790 AAUUUCAGUUGCAAAC-GCACUCCCGUU--UUGGCCUAUCCAUCAUUUUGGUUAGGCGCACAGUCGCGGUUACCAUAUUAGUGAAAUCUUGUAA-------------------------- .((((((..(((....-)))...((((.--.(((((((.(((......))).))))).)).....))))............)))))).......-------------------------- ( -20.60, z-score = -1.08, R) >droSec1.super_4 3278956 91 - 6179234 AAUUUCAGUUGCAAAC-GCACUCCCCUU--UUCGCCCAUCCAUUAUUUUGGUUAGGCGCACAGUCGCGGUUACCAUAUUAGUGCAAUCUUGUAA-------------------------- ........((((((..-(((((......--...(((...(((......)))...)))((......))............)))))....))))))-------------------------- ( -18.40, z-score = -1.21, R) >droYak2.chrX 17164110 119 - 21770863 AAUUUCAGUAGCAAAUUGCACUCCCCUU-UCUCGCCUAUCCAUCAUUUUGGUUAGGCGCACAGUCGCGGUUACAAUAUUAGAGACGCAUGGCACCGAAAUCUGAAUUGAAACUGUUGUAA ......(((.((.....)))))......-...((((((.(((......))).)))))).(((...((((((.((((.(((((..((........))...))))))))).))))))))).. ( -27.40, z-score = -0.75, R) >droEre2.scaffold_4690 1192840 110 + 18748788 UAUUUUAGUGGCAAACUGCACUCCCCCU-UCUCGCCCAUCCCUCAUUUUGGUUAGACGCACAGCCGCGGUUACAAUAUUAGUGACGCGUGAAACCGAAAUCUGUGCCACAA--------- .......((((((....((.........-....)).........(((((((((..((((.(......)(((((.......)))))))))..)))))))))...))))))..--------- ( -26.92, z-score = -1.49, R) >consensus AAUUUCAGUAGCAAAC_GCACUCCCCUU_UCUCGCCUAUCCAUCAUUUUGGUUAGGCGCACAGUCGCGGUUACCAUAUUAGUGAAAUCUUGUAA__________________________ .......(((((.....................(((((.(((......))).)))))((......)).)))))............................................... (-13.36 = -14.48 + 1.12)

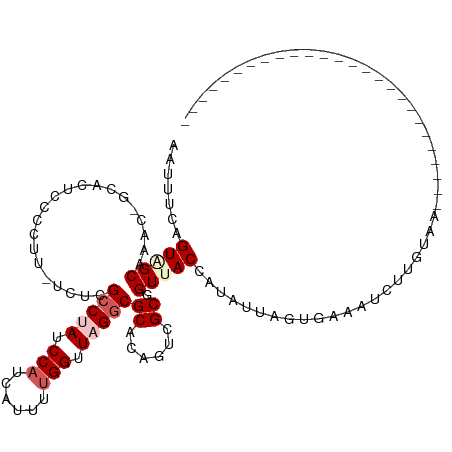

| Location | 3,808,494 – 3,808,587 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.90 |
| Shannon entropy | 0.43041 |
| G+C content | 0.44569 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -16.10 |
| Energy contribution | -15.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3808494 93 - 22422827 --------------------------UAACAAGAUUUCACUAAUAUGGUAACCGCGACUGUGCGCCUAACCAAAAUGAUGGAUAGGCGAGACAAGGGAAGUGC-GUUUGCUACCGAAAUU --------------------------......((((((.......((((((.((((.((((.((((((.(((......))).))))))..)).)).....)))-).))))))..)))))) ( -24.60, z-score = -2.18, R) >droSim1.chrX 2803973 91 - 17042790 --------------------------UUACAAGAUUUCACUAAUAUGGUAACCGCGACUGUGCGCCUAACCAAAAUGAUGGAUAGGCCAA--AACGGGAGUGC-GUUUGCAACUGAAAUU --------------------------......((((((((......)((((.((((.((((..(((((.(((......))).)))))...--.))))...)))-).))))...))))))) ( -25.80, z-score = -2.63, R) >droSec1.super_4 3278956 91 + 6179234 --------------------------UUACAAGAUUGCACUAAUAUGGUAACCGCGACUGUGCGCCUAACCAAAAUAAUGGAUGGGCGAA--AAGGGGAGUGC-GUUUGCAACUGAAAUU --------------------------.....((.(((((((.....)))((((((..((.(.((((((.(((......))).))))))..--.).))..))).-))).))))))...... ( -24.90, z-score = -1.99, R) >droYak2.chrX 17164110 119 + 21770863 UUACAACAGUUUCAAUUCAGAUUUCGGUGCCAUGCGUCUCUAAUAUUGUAACCGCGACUGUGCGCCUAACCAAAAUGAUGGAUAGGCGAGA-AAGGGGAGUGCAAUUUGCUACUGAAAUU ...................(((((((((((..((((.((((....((((....))))((...((((((.(((......))).))))))...-.)).))))))))....)).))))))))) ( -32.90, z-score = -1.52, R) >droEre2.scaffold_4690 1192840 110 - 18748788 ---------UUGUGGCACAGAUUUCGGUUUCACGCGUCACUAAUAUUGUAACCGCGGCUGUGCGUCUAACCAAAAUGAGGGAUGGGCGAGA-AGGGGGAGUGCAGUUUGCCACUAAAAUA ---------..((((((..((((.((.(..(.((((..((.......))...)))).((...((((((.((........)).))))))...-)).)..).)).))))))))))....... ( -31.00, z-score = -0.16, R) >consensus __________________________UUACAAGAUUUCACUAAUAUGGUAACCGCGACUGUGCGCCUAACCAAAAUGAUGGAUAGGCGAGA_AAGGGGAGUGC_GUUUGCAACUGAAAUU ...............................................((((.(((..((.(.((((((.(((......))).)))))).....).))..)))....)))).......... (-16.10 = -15.94 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:47 2011