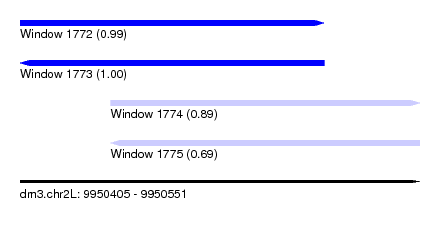
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,950,405 – 9,950,551 |
| Length | 146 |
| Max. P | 0.997912 |

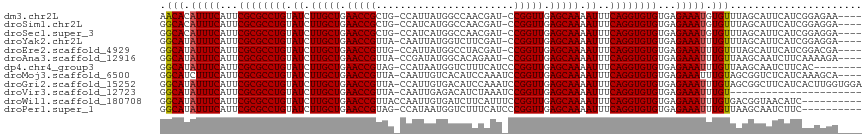
| Location | 9,950,405 – 9,950,516 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.19 |
| Shannon entropy | 0.38340 |
| G+C content | 0.43940 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -23.59 |
| Energy contribution | -23.68 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
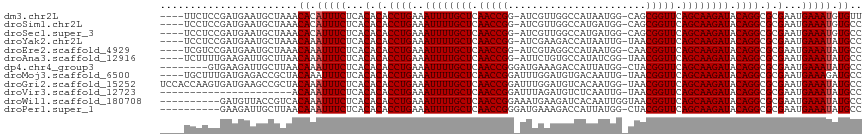
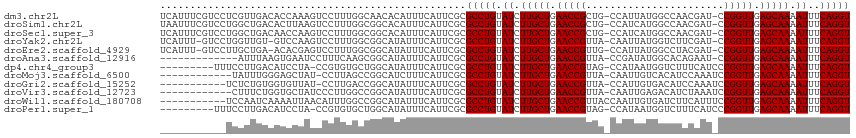
>dm3.chr2L 9950405 111 + 23011544 ----UUCUCCGAUGAAUGCUAAACACAUUUCUCACACACCUGAAAUUUUGCUCAACCGG-AUCGUUGGCCAUAAUGG-CAGCGGUUCAGCAAGAUACAGGCGCGAAUGAAAUGUGUU ----.................((((((((((...(.(.((((..((((((((.....((-(((((((.((.....))-))))))))))))))))).)))).).)...)))))))))) ( -37.80, z-score = -3.69, R) >droSim1.chr2L 9739848 111 + 22036055 ----UCCUCCGAUGAAUGCUAAACACAUUUCUCACACACCUGAAAUUUUGCUCAACCGG-AUCGUUGGCCAUGAUGG-CAGCGGUUCAGCAAGAUACAGGCGCGAAUGAAAUGUGCC ----...................((((((((...(.(.((((..((((((((.....((-(((((((.((.....))-))))))))))))))))).)))).).)...)))))))).. ( -35.80, z-score = -2.62, R) >droSec1.super_3 5397129 111 + 7220098 ----UCCUCCGAUGAAUGCUAAACACAUUUCUCACACACCUGAAAUUUUGCUCAACCGG-AUCGUUGGCCAUGAUGG-CAGCGGUUCAGCAAGAUACAGGCGCGAAUGAAAUGUGCC ----...................((((((((...(.(.((((..((((((((.....((-(((((((.((.....))-))))))))))))))))).)))).).)...)))))))).. ( -35.80, z-score = -2.62, R) >droYak2.chr2L 12640969 111 + 22324452 ----UCCUCCGAUGAAUGCUAAACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGG-AUCGAAGACCAUAAUUG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC ----...................((.(((((...(.(.((((..((((((((.(((((.-..(((.........)))-...))))).)))))))).)))).).)...))))).)).. ( -25.40, z-score = -2.07, R) >droEre2.scaffold_4929 10570533 111 + 26641161 ----UCGUCCGAUGAAUGCUAAACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGG-AUCGUAGGCCAUAAUGG-CAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC ----...................((.(((((...(.(.((((..((((((((.(((((.-.......(((.....))-)..))))).)))))))).)))).).)...))))).)).. ( -28.60, z-score = -1.60, R) >droAna3.scaffold_12916 13789760 111 + 16180835 ----UCUUUUGAAGAUUGCUUAACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGG-AUUCUGUGCCAUAUCGG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC ----...................((.(((((...(.(.((((..((((((((.(((((.-......((((.....))-)).))))).)))))))).)))).).)...))))).)).. ( -27.90, z-score = -1.98, R) >dp4.chr4_group3 11471775 108 - 11692001 --------GUGAAGAUUGCUUAACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGGGAUGAAAGACCAUUAUGG-CUACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC --------(((((((((........))))).)))).(.((((..((((((((.(((((.((((......))))....-...))))).)))))))).)))).)............... ( -26.20, z-score = -1.75, R) >droMoj3.scaffold_6500 14729856 112 + 32352404 ----UGCUUUGAUGAGACCGCUACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGGAUUUGGAUGUGACAAUUG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAGAUGCC ----..((((.((((((............)))).(.(.((((..((((((((.(((((...(((.......)))...-...))))).)))))))).)))).).).)).))))..... ( -26.70, z-score = -1.04, R) >droGri2.scaffold_15252 3661298 116 + 17193109 UCCACCAAGUGAUGAAGCCGCUACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGGAUUUGGAUGUCACAAUGG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC .......((((.......)))).((.(((((...(.(.((((..((((((((.(((((...(((.......)))...-...))))).)))))))).)))).).)...))))).)).. ( -27.80, z-score = -0.88, R) >droVir3.scaffold_12723 1805694 94 - 5802038 ----------------------ACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGGAUUUAGAUGUCUCAAUUG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC ----------------------.((.(((((...(.(.((((..((((((((.(((((......(((......))).-...))))).)))))))).)))).).)...))))).)).. ( -25.40, z-score = -3.07, R) >droWil1.scaffold_180708 12250841 107 + 12563649 ----------GAUGUUACCGUCACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGGAAAUGAAGAUCACAAUUGGUAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC ----------((((....)))).((.(((((...(.(.((((..((((((((.(((((...((.((........)).))..))))).)))))))).)))).).)...))))).)).. ( -27.60, z-score = -2.40, R) >droPer1.super_1 8587843 106 - 10282868 ----------GAAGAUUGCUUAACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGGGAUGAAAGACCAUUAUGG-CUACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC ----------.............((.(((((...(.(.((((..((((((((.(((((.((((......))))....-...))))).)))))))).)))).).)...))))).)).. ( -25.80, z-score = -2.10, R) >consensus ____UCCUCCGAUGAAUGCUAAACAAAUUUCUCACACACCUGAAAUUUUGCUCAACCGG_AUCGUAGGCCAUAAUGG_CAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCC .......................((.(((((...(.(.((((..((((((((.(((((.......................))))).)))))))).)))).).)...))))).)).. (-23.59 = -23.68 + 0.08)
| Location | 9,950,405 – 9,950,516 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.19 |
| Shannon entropy | 0.38340 |
| G+C content | 0.43940 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -30.97 |
| Energy contribution | -30.97 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.997912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
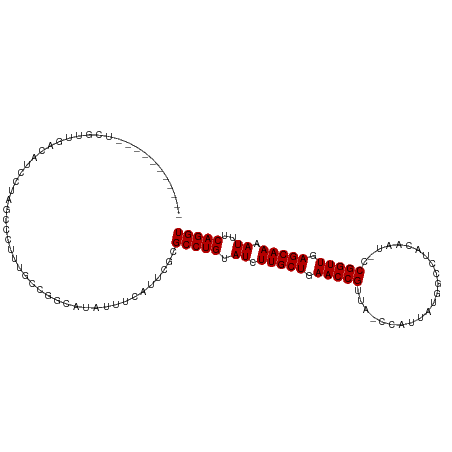
>dm3.chr2L 9950405 111 - 23011544 AACACAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGCUG-CCAUUAUGGCCAACGAU-CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUGUGUUUAGCAUUCAUCGGAGAA---- ((((((((((...((((((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))))))...)))))))))).................---- ( -40.90, z-score = -3.91, R) >droSim1.chr2L 9739848 111 - 22036055 GGCACAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGCUG-CCAUCAUGGCCAACGAU-CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUGUGUUUAGCAUUCAUCGGAGGA---- ((((((((((...((((((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))))))...)))))))))).................---- ( -40.80, z-score = -2.69, R) >droSec1.super_3 5397129 111 - 7220098 GGCACAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGCUG-CCAUCAUGGCCAACGAU-CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUGUGUUUAGCAUUCAUCGGAGGA---- ((((((((((...((((((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))))))...)))))))))).................---- ( -40.80, z-score = -2.69, R) >droYak2.chr2L 12640969 111 - 22324452 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CAAUUAUGGUCUUCGAU-CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUUUAGCAUUCAUCGGAGGA---- ((((.(((((...((((((((.((.(((((.(((((...-.......((((...)))-)))))).))))).))..))))))))...))))).)))).................---- ( -32.30, z-score = -1.78, R) >droEre2.scaffold_4929 10570533 111 - 26641161 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUG-CCAUUAUGGCCUACGAU-CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUUUAGCAUUCAUCGGACGA---- ((((.(((((...((((((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))))))...))))).)))).................---- ( -35.00, z-score = -2.11, R) >droAna3.scaffold_12916 13789760 111 - 16180835 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CCGAUAUGGCACAGAAU-CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUUAAGCAAUCUUCAAAAGA---- ((((.(((((...((((((((.((.(((((.(((((...-.................-.))))).))))).))..))))))))...))))).)))).................---- ( -31.90, z-score = -2.15, R) >dp4.chr4_group3 11471775 108 + 11692001 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUAG-CCAUAAUGGUCUUUCAUCCCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUUAAGCAAUCUUCAC-------- ((((.(((((...((((((((.((.(((((.(((((.((-((.....)).)).......))))).))))).))..))))))))...))))).)))).............-------- ( -34.10, z-score = -3.71, R) >droMoj3.scaffold_6500 14729856 112 - 32352404 GGCAUCUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CAAUUGUCACAUCCAAAUCCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUAGCGGUCUCAUCAAAGCA---- .(((..((((...((((((((.((.(((((.(((((...-...................))))).))))).))..))))))))...))))..))).((.(......)...)).---- ( -31.65, z-score = -2.21, R) >droGri2.scaffold_15252 3661298 116 - 17193109 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CCAUUGUGACAUCCAAAUCCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUAGCGGCUUCAUCACUUGGUGGA .(((.(((((...((((((((.((.(((((.((((((((-(....)))))..........)))).))))).))..))))))))...))))).)))......((((((....)))))) ( -36.30, z-score = -2.07, R) >droVir3.scaffold_12723 1805694 94 + 5802038 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CAAUUGAGACAUCUAAAUCCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGU---------------------- .(((.(((((...((((((((.((.(((((.(((((...-.....((....))......))))).))))).))..))))))))...))))).)))---------------------- ( -32.14, z-score = -4.17, R) >droWil1.scaffold_180708 12250841 107 - 12563649 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUACCAAUUGUGAUCUUCAUUUCCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUGACGGUAACAUC---------- .(((.(((((...((((((((.((.(((((.(((((..(......(((.....))))..))))).))))).))..))))))))...))))).)))............---------- ( -32.70, z-score = -2.83, R) >droPer1.super_1 8587843 106 + 10282868 GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUAG-CCAUAAUGGUCUUUCAUCCCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUUAAGCAAUCUUC---------- ((((.(((((...((((((((.((.(((((.(((((.((-((.....)).)).......))))).))))).))..))))))))...))))).))))...........---------- ( -34.10, z-score = -3.93, R) >consensus GGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA_CCAUUAUGGCCUACAAU_CCGGUUGAGCAAAAUUUCAGGUGUGUGAGAAAUUUGUUUAGCAUUCAUCGGAGGA____ .(((.(((((...((((((((.((.(((((.(((((.......................))))).))))).))..))))))))...))))).)))...................... (-30.97 = -30.97 + 0.01)
| Location | 9,950,438 – 9,950,551 |
|---|---|
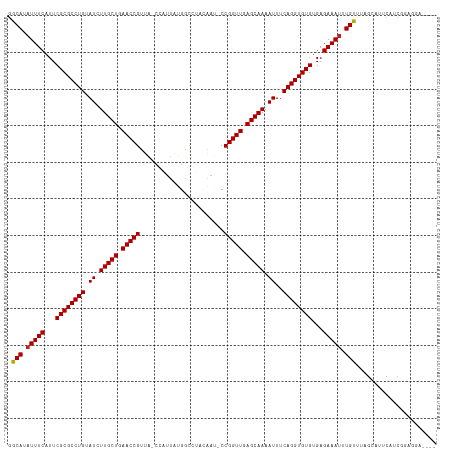
| Length | 113 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Shannon entropy | 0.52968 |
| G+C content | 0.45992 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -18.55 |
| Energy contribution | -18.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.892680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9950438 113 + 23011544 ACCUGAAAUUUUGCUCAACCGG-AUCGUUGGCCAUAAUGG-CAGCGGUUCAGCAAGAUACAGGCGCGAAUGAAAUGUGUUGCCAAAGGACUUUGGUGUCAACGAGGACGAAAUGA .((((..((((((((.....((-(((((((.((.....))-))))))))))))))))).)))).................((((((....))))))(((......)))....... ( -32.60, z-score = -0.73, R) >droSim1.chr2L 9739881 113 + 22036055 ACCUGAAAUUUUGCUCAACCGG-AUCGUUGGCCAUGAUGG-CAGCGGUUCAGCAAGAUACAGGCGCGAAUGAAAUGUGCCGCCAAAGGACUUAAGUGUCAGCCAGGACGAAAUUA .((((..((((((((.....((-(((((((.((.....))-)))))))))))))))))...((((((.......)))))).......(((......)))...))))......... ( -36.10, z-score = -1.48, R) >droSec1.super_3 5397162 113 + 7220098 ACCUGAAAUUUUGCUCAACCGG-AUCGUUGGCCAUGAUGG-CAGCGGUUCAGCAAGAUACAGGCGCGAAUGAAAUGUGCCGCCAAAGGACUUGGUUGUCAGCCAGGACGAAAUGA .(((...((((((((.....((-(((((((.((.....))-)))))))))))))))))...((((((.......)))))).....))).((((((.....))))))......... ( -37.80, z-score = -1.31, R) >droYak2.chr2L 12641002 111 + 22324452 ACCUGAAAUUUUGCUCAACCGG-AUCGAAGACCAUAAUUG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGCCAAAGGACUUGGAC-ACAACCAGGAC-AAAUGA .(((...((((((((.(((((.-..(((.........)))-...))))).))))))))...((((((.........)).))))..))).(((((..-....)))))..-...... ( -29.20, z-score = -2.46, R) >droEre2.scaffold_4929 10570566 111 + 26641161 ACCUGAAAUUUUGCUCAACCGG-AUCGUAGGCCAUAAUGG-CAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGCCAAAGGACUCGUGU-UCAGCAAGGAC-AAAUGA .(((...((((((((.(((((.-.......(((.....))-)..))))).))))))))...((((((.........)).))))..))).(...(((-((.....))))-)...). ( -30.70, z-score = -0.95, R) >droAna3.scaffold_12916 13789793 100 + 16180835 ACCUGAAAUUUUGCUCAACCGG-AUUCUGUGCCAUAUCGG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGCUUGAAAGGAUUCACUUAAAU------------- .(((...((((((((.(((((.-......((((.....))-)).))))).)))))))).((((((((.........)).))))))..)))............------------- ( -27.50, z-score = -1.92, R) >dp4.chr4_group3 11471804 104 - 11692001 ACCUGAAAUUUUGCUCAACCGGGAUGAAAGACCAUUAUGG-CUACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCAGCACACGG-UAGGAUGUCAAGGAAA--------- .((((..((((((((.(((((.((((......))))....-...))))).)))))))).))))......(((....((((.......))-)).....)))......--------- ( -26.00, z-score = -0.90, R) >droMoj3.scaffold_6500 14729889 101 + 32352404 ACCUGAAAUUUUGCUCAACCGGAUUUGGAUGUGACAAUUG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAGAUGCCGGCUAAGG-AUAGCUCCCAAAUA------------ .((((..((((((((.(((((...(((.......)))...-...))))).)))))))).)))).............((..(((((...-.)))))..))....------------ ( -24.90, z-score = -0.71, R) >droGri2.scaffold_15252 3661335 102 + 17193109 ACCUGAAAUUUUGCUCAACCGGAUUUGGAUGUCACAAUGG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGGUCAAGG-AUAACACCACAGAGA----------- .(((((.((((((((.(((((...(((.......)))...-...))))).))))))))...((((...........))))..)).)))-...............----------- ( -22.90, z-score = -0.14, R) >droVir3.scaffold_12723 1805709 102 - 5802038 ACCUGAAAUUUUGCUCAACCGGAUUUAGAUGUCUCAAUUG-UAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGGCCAAGGGAUAGCACCAGAAAG------------ .((((..((((((((.(((((......(((......))).-...))))).)))))))).))))......((...(((.((......)).))).))........------------ ( -24.90, z-score = -0.90, R) >droWil1.scaffold_180708 12250868 104 + 12563649 ACCUGAAAUUUUGCUCAACCGGAAAUGAAGAUCACAAUUGGUAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGGCCAAAUGUUAAUUUUGAUUGGA----------- .((((..((((((((.(((((...((.((........)).))..))))).)))))))).))))...............(((..((((.......))))..))).----------- ( -24.80, z-score = -0.97, R) >droPer1.super_1 8587870 104 - 10282868 ACCUGAAAUUUUGCUCAACCGGGAUGAAAGACCAUUAUGG-CUACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCAGCACACGG-UAGGAUGUCAAGGAAA--------- .((((..((((((((.(((((.((((......))))....-...))))).)))))))).))))......(((....((((.......))-)).....)))......--------- ( -26.00, z-score = -0.90, R) >consensus ACCUGAAAUUUUGCUCAACCGG_AUCGUAGGCCAUAAUGG_CAACGGUUCAGCAAGAUACAGGCGCGAAUGAAAUAUGCCGCCAAAGGACUAGGAUGUCAACGA___________ .((((..((((((((.(((((.......................))))).)))))))).)))).................................................... (-18.55 = -18.55 + 0.00)

| Location | 9,950,438 – 9,950,551 |
|---|---|
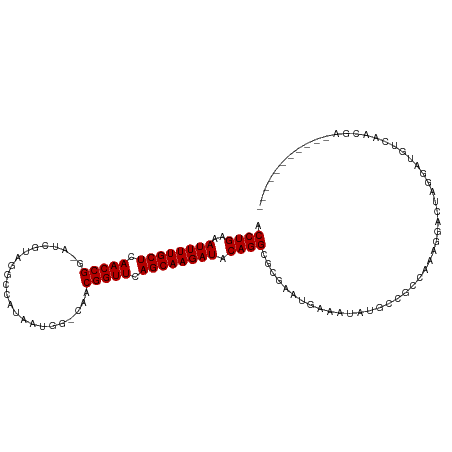
| Length | 113 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Shannon entropy | 0.52968 |
| G+C content | 0.45992 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -18.42 |
| Energy contribution | -18.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.56 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9950438 113 - 23011544 UCAUUUCGUCCUCGUUGACACCAAAGUCCUUUGGCAACACAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGCUG-CCAUUAUGGCCAACGAU-CCGGUUGAGCAAAAUUUCAGGU .............((((...(((((....))))))))).............(((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))) ( -29.10, z-score = -1.41, R) >droSim1.chr2L 9739881 113 - 22036055 UAAUUUCGUCCUGGCUGACACUUAAGUCCUUUGGCGGCACAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGCUG-CCAUCAUGGCCAACGAU-CCGGUUGAGCAAAAUUUCAGGU .......((((..(..(((......)))..)..).))).............(((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))) ( -30.20, z-score = -0.38, R) >droSec1.super_3 5397162 113 - 7220098 UCAUUUCGUCCUGGCUGACAACCAAGUCCUUUGGCGGCACAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGCUG-CCAUCAUGGCCAACGAU-CCGGUUGAGCAAAAUUUCAGGU .......((((..(..(((......)))..)..).))).............(((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))) ( -30.20, z-score = -0.09, R) >droYak2.chr2L 12641002 111 - 22324452 UCAUUU-GUCCUGGUUGU-GUCCAAGUCCUUUGGCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CAAUUAUGGUCUUCGAU-CCGGUUGAGCAAAAUUUCAGGU .....(-(((.(((....-..))).(((....)))))))............(((((.((.(((((.(((((...-.......((((...)))-)))))).))))).))..))))) ( -25.50, z-score = -0.14, R) >droEre2.scaffold_4929 10570566 111 - 26641161 UCAUUU-GUCCUUGCUGA-ACACGAGUCCUUUGGCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUG-CCAUUAUGGCCUACGAU-CCGGUUGAGCAAAAUUUCAGGU .((...-(..((((....-...))))..)..))((((..........))))(((((.((.(((((.(((((..(-((.....))).......-.))))).))))).))..))))) ( -27.90, z-score = -0.15, R) >droAna3.scaffold_12916 13789793 100 - 16180835 -------------AUUUAAGUGAAUCCUUUCAAGCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CCGAUAUGGCACAGAAU-CCGGUUGAGCAAAAUUUCAGGU -------------.......((((....)))).((((..........))))(((((.((.(((((.(((((...-.................-.))))).))))).))..))))) ( -22.90, z-score = -0.41, R) >dp4.chr4_group3 11471804 104 + 11692001 ---------UUUCCUUGACAUCCUA-CCGUGUGCUGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUAG-CCAUAAUGGUCUUUCAUCCCGGUUGAGCAAAAUUUCAGGU ---------....(((((.......-....((((.(((.............))).)))).(((((.(((((.((-((.....)).)).......))))).)))))....))))). ( -23.42, z-score = -0.56, R) >droMoj3.scaffold_6500 14729889 101 - 32352404 ------------UAUUUGGGAGCUAU-CCUUAGCCGGCAUCUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CAAUUGUCACAUCCAAAUCCGGUUGAGCAAAAUUUCAGGU ------------....((..((....-((......))...))..)).....(((((.((.(((((.(((((...-...................))))).))))).))..))))) ( -23.15, z-score = -0.63, R) >droGri2.scaffold_15252 3661335 102 - 17193109 -----------UCUCUGUGGUGUUAU-CCUUGACCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CCAUUGUGACAUCCAAAUCCGGUUGAGCAAAAUUUCAGGU -----------....(((.(.((((.-...))))).)))............(((((.((.(((((.((((((((-(....)))))..........)))).))))).))..))))) ( -24.80, z-score = -1.02, R) >droVir3.scaffold_12723 1805709 102 + 5802038 ------------CUUUCUGGUGCUAUCCCUUGGCCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA-CAAUUGAGACAUCUAAAUCCGGUUGAGCAAAAUUUCAGGU ------------.......(((((..((...))..)))))...........(((((.((.(((((.(((((...-.....((....))......))))).))))).))..))))) ( -24.94, z-score = -0.84, R) >droWil1.scaffold_180708 12250868 104 - 12563649 -----------UCCAAUCAAAAUUAACAUUUGGCCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUACCAAUUGUGAUCUUCAUUUCCGGUUGAGCAAAAUUUCAGGU -----------.((..(((((.......)))))..))..............(((((.((.(((((.(((((..(......(((.....))))..))))).))))).))..))))) ( -21.00, z-score = -0.55, R) >droPer1.super_1 8587870 104 + 10282868 ---------UUUCCUUGACAUCCUA-CCGUGUGCUGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUAG-CCAUAAUGGUCUUUCAUCCCGGUUGAGCAAAAUUUCAGGU ---------....(((((.......-....((((.(((.............))).)))).(((((.(((((.((-((.....)).)).......))))).)))))....))))). ( -23.42, z-score = -0.56, R) >consensus ___________UCGUUGACAUCCUAGCCCUUUGCCGGCAUAUUUCAUUCGCGCCUGUAUCUUGCUGAACCGUUA_CCAUUAUGGCCUACAAU_CCGGUUGAGCAAAAUUUCAGGU ...................................................(((((.((.(((((.(((((.......................))))).))))).))..))))) (-18.42 = -18.43 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:19 2011