
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,780,292 – 3,780,412 |
| Length | 120 |
| Max. P | 0.609275 |

| Location | 3,780,292 – 3,780,412 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.62 |
| Shannon entropy | 0.15400 |
| G+C content | 0.39880 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.38 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.577741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
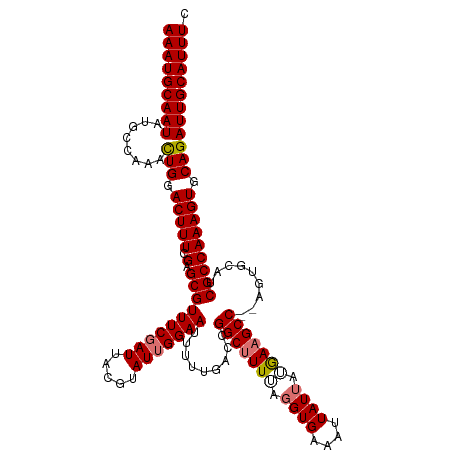
>dm3.chrX 3780292 120 + 22422827 AAAUGCAAUAUGCCAAACUGGACUUUCGAGCGUUUCGAUAACGUAUUGGAAUUUUCGAUCGGCUUUAAGGUGAAAUUAUUAUAAAGCCAAAGUGCAUCGCCAAAGUGCAGAUUGCAUUUC (((((((((.(((.....(((....(((((.(((((((((...))))))))).)))))..(((((((..(((....)))..)))))))...........)))....))).))))))))). ( -34.80, z-score = -2.78, R) >droEre2.scaffold_4690 1164056 118 + 18748788 AAAUGCAAUAUGCCACAUUGGACUUUCGAGCGUUUCCAUUACGUAUUGGAAUUUUUGACCGGCAUUUUGGUGAAAUUAUAAUGAAGCC--AGUGCAGCGCCAAAGUGCAGAUUGCAUUUU (((((((((.(((.((.((((....(((((...(((((........)))))..)))))...((((..((((...((....))...)))--)))))....)))).))))).))))))))). ( -32.70, z-score = -1.61, R) >droYak2.chrX 17136274 118 - 21770863 AAAUGCAAUAUGCCAAACUGAACUUUCGAGCGUUUCGAUUACGUAUUGGAAUUUUUGACCGGCUUUUCGGUGAAAUUAUUAAGAAGCC--AGUGCAACGCCAAAGUGCAGAUUGCAUUUC (((((((((.(((........(((((.(.((((((((((.....)))))))...(((((.(((((((..(((....)))..)))))))--.)).))))))))))))))).))))))))). ( -32.70, z-score = -2.15, R) >droSec1.super_4 3251750 118 - 6179234 AAAUGCAAUAUGGCAAACUGGACUUUCGAGCGUUUCGAUUACGUAUUGGAAUUUGGGAUCGGCUUUUAGGUGAAAUUAUUAUGAAGCC--AGUGCGUCGCCAAAGUGCAGAUUGCAUUUC (((((((((...(((...(((....(((((...)))))..(((((((((.((.(((..(((.(.....).)))..)))..))....))--)))))))..)))...)))..))))))))). ( -31.30, z-score = -0.91, R) >consensus AAAUGCAAUAUGCCAAACUGGACUUUCGAGCGUUUCGAUUACGUAUUGGAAUUUUUGACCGGCUUUUAGGUGAAAUUAUUAUGAAGCC__AGUGCAUCGCCAAAGUGCAGAUUGCAUUUC (((((((((........(((.(((((.(.((((((((((.....))))))).........(((((((.((((....)))).))))))).........))))))))).)))))))))))). (-25.62 = -26.38 + 0.75)


| Location | 3,780,292 – 3,780,412 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.62 |
| Shannon entropy | 0.15400 |
| G+C content | 0.39880 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3780292 120 - 22422827 GAAAUGCAAUCUGCACUUUGGCGAUGCACUUUGGCUUUAUAAUAAUUUCACCUUAAAGCCGAUCGAAAAUUCCAAUACGUUAUCGAAACGCUCGAAAGUCCAGUUUGGCAUAUUGCAUUU .(((((((((.(((...(((((((.((...(((((((((..............)))))))))((((.(((........))).))))...))))).....))))....))).))))))))) ( -32.54, z-score = -3.17, R) >droEre2.scaffold_4690 1164056 118 - 18748788 AAAAUGCAAUCUGCACUUUGGCGCUGCACU--GGCUUCAUUAUAAUUUCACCAAAAUGCCGGUCAAAAAUUCCAAUACGUAAUGGAAACGCUCGAAAGUCCAAUGUGGCAUAUUGCAUUU .(((((((((.(((((.((((.((((.(((--(((......................))))))))....(((((........)))))..)).(....).)))).)).))).))))))))) ( -31.95, z-score = -2.53, R) >droYak2.chrX 17136274 118 + 21770863 GAAAUGCAAUCUGCACUUUGGCGUUGCACU--GGCUUCUUAAUAAUUUCACCGAAAAGCCGGUCAAAAAUUCCAAUACGUAAUCGAAACGCUCGAAAGUUCAGUUUGGCAUAUUGCAUUU .(((((((((.(((...((((..(((.(((--(((((.((............)).))))))))))).....))))..........((((...(....)....)))).))).))))))))) ( -30.10, z-score = -2.08, R) >droSec1.super_4 3251750 118 + 6179234 GAAAUGCAAUCUGCACUUUGGCGACGCACU--GGCUUCAUAAUAAUUUCACCUAAAAGCCGAUCCCAAAUUCCAAUACGUAAUCGAAACGCUCGAAAGUCCAGUUUGCCAUAUUGCAUUU .(((((((((..(((..(((((.(((....--(((((..................)))))((........)).....)))..((((.....))))..).))))..)))...))))))))) ( -24.07, z-score = -1.40, R) >consensus GAAAUGCAAUCUGCACUUUGGCGAUGCACU__GGCUUCAUAAUAAUUUCACCUAAAAGCCGAUCAAAAAUUCCAAUACGUAAUCGAAACGCUCGAAAGUCCAGUUUGGCAUAUUGCAUUU .(((((((((.(((...(((((..........(((((..................)))))......................((((.....))))..).))))....))).))))))))) (-20.40 = -20.77 + 0.37)



Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:39 2011