| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,779,395 – 3,779,495 |
| Length | 100 |
| Max. P | 0.957060 |

| Location | 3,779,395 – 3,779,495 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.51 |
| Shannon entropy | 0.34774 |
| G+C content | 0.45543 |
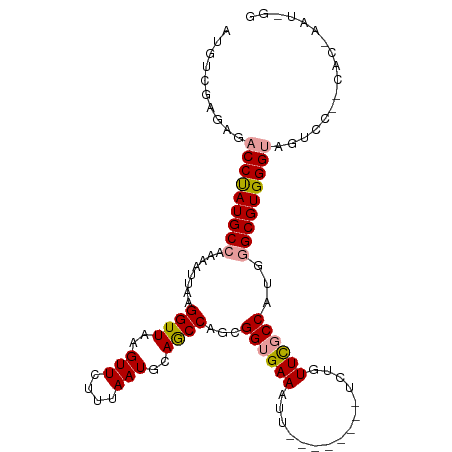
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -20.71 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
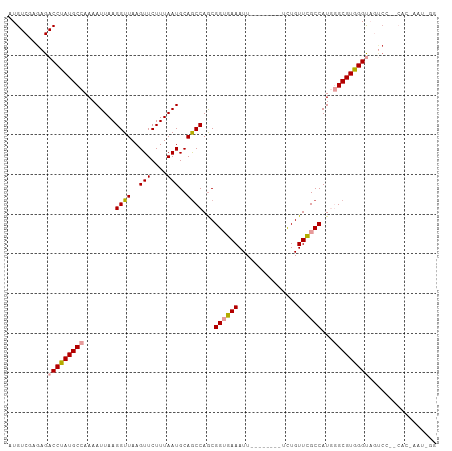
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
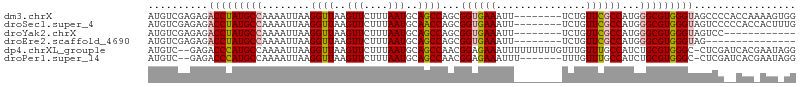
>dm3.chrX 3779395 100 - 22422827 AUGUCGAGAGACCUAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAGCCAGCGGUGAAAUU--------UCUGUUCGCCAUGGGCGUGGGUAGCCCCACCAAAAGUGG .......(..(((((((((...(((((((.......)))))))((.....))((((((...--------....))))))...)))))))))..).((((.....)))) ( -30.30, z-score = -1.53, R) >droSec1.super_4 3250810 100 + 6179234 AUGUCGAGAGACCUAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAACCAGCGGUGAAAUU--------UCUGUUCGCCAUGGGCGUGGGUAGUCCCCCACCACUUUG ..(((....)))....(((...(((((((.......)))))))((.....))((((((...--------....))))))...)))(((((......)))))....... ( -27.60, z-score = -1.31, R) >droYak2.chrX 17135382 88 + 21770863 AUGUCGAGAGACCUAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAGCCAGCGGUGAAAUU--------UCUGUUCGCCAUGGGCGUGGGUAGUCC------------ .......((.(((((((((...(((((((.......)))))))((.....))((((((...--------....))))))...)))))))))..)).------------ ( -26.80, z-score = -2.01, R) >droEre2.scaffold_4690 1163223 85 - 18748788 AUGUCGAGAGACCUAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAGCCAGCGGUGAAAUU--------UCUGUUCGCCAUGGGCGUGGGUAG--------------- ..........(((((((((...(((((((.......)))))))((.....))((((((...--------....))))))...)))))))))..--------------- ( -26.30, z-score = -2.04, R) >dp4.chrXL_group1e 626585 105 + 12523060 AUGUC--GAGACCCAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAGCCAACGGAGAAAUUUUUUUUUGUUUGUUUGCCAUCUGCGUGGGC-CUCGAUCACGAAUAGG ..(((--(((.(((((((....(((((((.......)))))))(((((.(((((((((....))))))))).).)))).....))))))).-)))))).......... ( -32.50, z-score = -2.93, R) >droPer1.super_14 1716193 98 - 2168203 AUGUC--GAGACCCAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAGCCAACGGAGAAAUUU-------UUUGUUUGCCAUCUGCGUGGGC-CUCGAUCACGAAUAGG ..(((--(((.(((((((....(((((((.......)))))))((((.(((..((....)).-------.))).)))).....))))))).-)))))).......... ( -31.10, z-score = -2.91, R) >consensus AUGUCGAGAGACCUAUGCCAAAAUUAAGGUUAAGUUCUUUAAUGCAGCCAGCGGUGAAAUU________UCUGUUCGCCAUGGGCGUGGGUAGUCC__CAC_AAU_GG ..........(((((((((........((((..(((....)))..))))...((((((...............))))))...)))))))))................. (-20.71 = -21.13 + 0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:37 2011