| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,777,256 – 3,777,357 |
| Length | 101 |
| Max. P | 0.629625 |

| Location | 3,777,256 – 3,777,357 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 77.34 |
| Shannon entropy | 0.42955 |
| G+C content | 0.30427 |
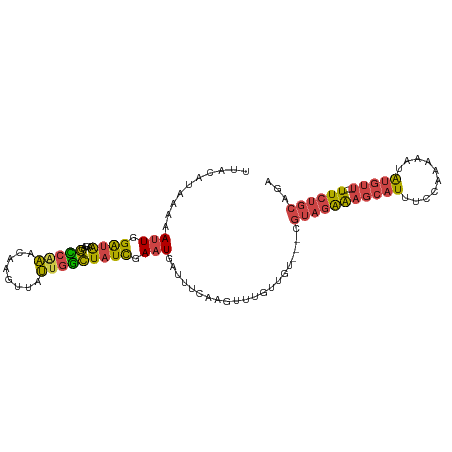
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -8.39 |
| Energy contribution | -8.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.629625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
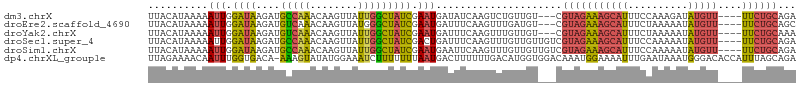
>dm3.chrX 3777256 101 + 22422827 UUACAUAAAAAUUGGAUAAGAUGCCAAACAAGUUAUUGGCUAUCGAAUGAUAUCAAGUCUGUUGU---CGUAGAAAGCAUUUCCAAAGAUAUGUU----UUCUGCAGA ..............(((((((((((((........))))).)))....(((.....)))..))))---)(((((((((((.((....)).)))))----))))))... ( -25.70, z-score = -2.94, R) >droEre2.scaffold_4690 1161155 101 + 18748788 UUACAUAAAAAUUGGAUAAGAUGUCAAACAAGUUAUGGGCUAUCGAAUGAUUUCAAGUUUGAUGU---CGUAGAAAGCAUUUCUAAAAAUAUGUU----UUCUGCAGC ...................((((((((((.(((.....)))...(((....)))..)))))))))---)(((((((((((..........)))))----))))))... ( -25.90, z-score = -3.39, R) >droYak2.chrX 17133038 101 - 21770863 UUACAUAAAAAUUGGAUAAGAUGUCAAACAAGUUAUUGGCUAUCGAAUGAUUUCAAGUUUGUUGU---CGUAGAAAGCAUUUCUAAAAAUAUGUU----UUCUGCAAA ...................((((.(((((((((((((.(....).))))))))...))))).)))---)(((((((((((..........)))))----))))))... ( -22.20, z-score = -2.44, R) >droSec1.super_4 3248713 104 - 6179234 UUACAUAAAAAUUGGAUAAGAUGCCAAACAAGUUAUUGGCUAUCGACUGAUUUCAAGUUUGUUGUUGUCGUAGAAAGCAUUUCCAAAAAUAUGUU----UUCUGCAGA ..............(((((((((((((........))))).)))((((.......)))).....)))))(((((((((((..........)))))----))))))... ( -24.80, z-score = -2.34, R) >droSim1.chrX 2783133 104 + 17042790 UUACAUAAAAAUUGGAUAAGAUGCCAAACAAGUUAUUGGCUAUCGAAUGAAUUCAAGUUUGUUGUUGUCGUAGAAAGCAUUUCCAAAAAUAUGUU----UUCUGCAGA ..............(((((((((((((........))))).))).((..(((....)))..)).)))))(((((((((((..........)))))----))))))... ( -25.10, z-score = -2.73, R) >dp4.chrXL_group1e 624714 107 - 12523060 UUAGAAAACAAUUUGGUGACA-AAAGUAUAUGGAAAUCUUUUUUUAAUGACUUUUUUGACAUGGUGGACAAAUGGAAAAUUUGAAUAAAUGGGACACCAUUUAGCAGA (((((((....((((....))-))(((...((((((....))))))...))))))))))....((...(((((.....)))))..(((((((....)))))))))... ( -17.70, z-score = -0.68, R) >consensus UUACAUAAAAAUUGGAUAAGAUGCCAAACAAGUUAUUGGCUAUCGAAUGAUUUCAAGUUUGUUGU___CGUAGAAAGCAUUUCCAAAAAUAUGUU____UUCUGCAGA ..........(((.((((....(((((........))))))))).))).....................(((((((((((..........)))))....))))))... ( -8.39 = -8.45 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:35 2011