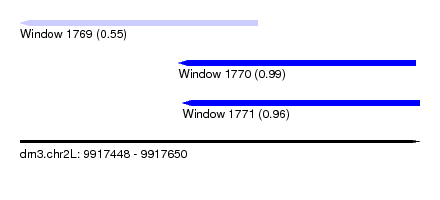
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,917,448 – 9,917,650 |
| Length | 202 |
| Max. P | 0.988704 |

| Location | 9,917,448 – 9,917,568 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.50 |
| Shannon entropy | 0.61523 |
| G+C content | 0.52240 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -7.36 |
| Energy contribution | -7.10 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.91 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.545528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
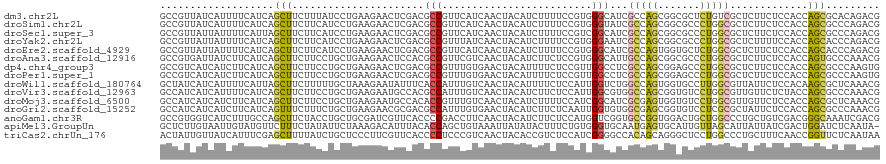
>dm3.chr2L 9917448 120 - 23011544 GCCGUUAUCAUUUUCAUCAGCUUCUUUAUCCUGAAGAACUCGACGCCGUUCAUCAACUACAUCUUUUCCGUGGGCAUCGCCAGCGGCGCUCUGUCGCUCUUCUCCACCAGCGCACAGACG ((((((......(((.((((..........)))).)))..(((.((((((....)))..(((.......)))))).)))..))))))(.(((((((((..........)))).)))))). ( -28.90, z-score = -1.74, R) >droSim1.chr2L 9707017 120 - 22036055 GCCGUUAUCAUUUUCAUCAGCUUCUUCAUCCUGAAGAACUCGACGCCGUUCAUCAACUACAUCUUUUCCGUGGGUAUCGCCAGCGGCGCCCUGGCGCUCUUCUCCACCAGCGCCCAGACG ((((((............((.((((((.....))))))))(((..(((((....)))..(((.......)))))..)))..))))))...((((((((..........)))).))))... ( -30.70, z-score = -1.70, R) >droSec1.super_3 5364993 120 - 7220098 GCCGUUAUUAUUUUCAUUAGCUUCUUCAUCCUGAAGAACUCGACGCCGUUCAUCAACUACAUCUUUUCCGUCGGCAUCGCCAGCGGCGCCCUGGCGCUCUUCUCCACCAGCGCCCAGACG ((((((............((.((((((.....))))))))(((.((((.......................)))).)))..))))))...((((((((..........)))).))))... ( -32.40, z-score = -2.88, R) >droYak2.chr2L 12608673 120 - 22324452 GCCGUUAUUAUUUUCAUCAGCUUCUUCAUCCUGAAGAACUCGACGCCGUUUAUCAACUACAUCUUUUCCGUGGGAAUCGCCAGCGGCGCCCUGGCGCUCUUUUCCACCAGCACCCAGACG ..((((..........((((.((((((.....)))))))).)).((.(((....)))............(((((((.((((((.(....)))))))....)))))))..)).....)))) ( -28.60, z-score = -1.62, R) >droEre2.scaffold_4929 10533471 120 - 26641161 GCCGUUAUUAUUUUCAUCAGCUUCUUCAUCCUGAAGAACUCGACGCCGUUCAUCAACUACAUCUUUUCCGUGGGCAUCGCCAGUGGUGCUCUGGCGCUCUUCUCCACCAGCACCCAGACG ..((((............((.((((((.....)))))))).(((((((((....)))..............((((((((....)))))))).)))).)).................)))) ( -25.50, z-score = -0.50, R) >droAna3.scaffold_12916 13757188 120 - 16180835 GCCGUGAUUAUCUUCAUCAGCUUCUUCCUGCUGAAGAACUCCACGCCGUUCGUCAACUACAUCUUCUCCGUGGGCAUUGCCAGCGGCGCCCUGGCGCUCUUCUCCACCAGUGCCCAAACG (.((((....((((((.(((.......))).))))))....)))).).......................(((((((((..((.(((((....)))))...))....))))))))).... ( -35.20, z-score = -2.65, R) >dp4.chr4_group3 11438479 120 + 11692001 GCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAACUCGACGCCGUUUGUGAACUACAUUUUCUCCGUUGGCCUCGCCAGCGGAGCCCUGGCGCUCUUCUCCACCAGCGCCCAAGUG (.((((....((((((.(((.......))).))))))....)))).)............(((((.((((((((((...))))))))))....((((((..........)))))).))))) ( -39.80, z-score = -3.53, R) >droPer1.super_1 8554616 120 + 10282868 GCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAACUCGACGCCGUUUGUGAACUACAUUUUCUCCGUUGGCCUCGCCAGCGGAGCCCUGGCGCUCUUCUCCACCAGCGCCCAAGUG (.((((....((((((.(((.......))).))))))....)))).)............(((((.((((((((((...))))))))))....((((((..........)))))).))))) ( -39.80, z-score = -3.53, R) >droWil1.scaffold_180764 2526014 120 - 3949147 GCUAUCAUCAUUUUCAUUAGCUUCUUUUUGCUAAAGAAUAUUUCACCAUUUGUCAACUACAUUUUCUCCAUUGGUCUGGCCAGUGGUGCCUUGGCGUUAUUCUCCACAAGCGCUCAAACG ............(((.(((((........))))).))).....(((((((.((((((((............)))).)))).)))))))..((((((((..........))))).)))... ( -25.50, z-score = -2.55, R) >droVir3.scaffold_12963 14554921 120 - 20206255 GCCAUCAUCAUUUUCAUCAGCUUCUUCCUGCUGAAGAAUGCCACGCCAUUUGUCAACUACAUCUUCUCCAUUGGCGUGGCCAGCGGUGUCCUGGCGUUGUUCUCUACCAGCGCCCAAACG (((.........(((.(((((........))))).))).(((((((((.......................)))))))))....)))((...(((((((........)))))))...)). ( -39.90, z-score = -4.50, R) >droMoj3.scaffold_6500 17474746 120 - 32352404 GCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAAUGCCACACCGUUUGUCAACUACAUCUUUUCCAUCGGCAUCGCGAGUGGUGUCCUGGCGUUGUUCUCCACCAGCGCUCAAACG ............(((.(((((........))))).)))........((((((....................(((((((....)))))))..(((((((........))))))))))))) ( -29.40, z-score = -1.73, R) >droGri2.scaffold_15252 8251262 120 - 17193109 GCCAUCAUCAUCUUCAUCAGUUUCUUUCUGCUGAAGAACGCGACGCCAUUUGUGAACUACAUCUUCUCAAUUGGUGUGGCGAGCGGUGUCCUCGCGCUAUUCUCCACCAGCGCCCAAACG (((.......((((((.(((.......))).)))))).(((.((((((.(((.(((.......))).))).)))))).)))...)))......(((((..........)))))....... ( -38.40, z-score = -3.57, R) >anoGam1.chr3R 35788377 120 + 53272125 GCCGUGGUCAUCUUUGCCAGCUUCUACCUGCUGCGAUCGUUCACCCCGACCUUCAACUACAUCUUCUCCAUGGUCGGUGCCGGUGGACUGCUGGCCCUGCUGUCGACGGGCAAAUCGACG .((((.(.((.(...((((((.((((((.((...((....))...((((((....................)))))).)).))))))..))))))...).)).).))))........... ( -42.75, z-score = -2.32, R) >apiMel3.GroupUn 257803994 119 + 399230636 GCUCUUGUAAUUGUAUGUUCUUUCUAUAUUCUAAAGACAUUUACACCAGCUGUAAAUUAUAUACUUUCUGUGGGUGCAAUGAGUGCAUUGUUAGCAUUAUUAUCGACUGGAUCUCAAUA- ((.((..((...(((((((((((.........))))).(((((((.....))))))).))))))....))..)).))..(((((.(((((.(((.....))).))).)).).))))...- ( -17.70, z-score = 1.30, R) >triCas2.chrUn_176 67351 120 + 68640 ACUAUUGUUAUCAUUUCGAGCUUUUAUCUGCUCCCUUCGUUCACCCCUCCCGUCAACUACACCGUCUCCAUCGGGGCCACAGCAGGGCUCCUGGCCCUGCUUUCAACCGGUUCUCAAUAA ..(((((..(((.....((((........)))).....((...((((........((......)).......))))..))((((((((.....)))))))).......)))...))))). ( -30.46, z-score = -2.83, R) >consensus GCCGUUAUCAUUUUCAUCAGCUUCUUCCUGCUGAAGAACUCGACGCCGUUUGUCAACUACAUCUUCUCCGUGGGCAUCGCCAGCGGCGCCCUGGCGCUCUUCUCCACCAGCGCCCAAACG ...................(((......................(((.........................)))...(((((.......))))).............)))......... ( -7.36 = -7.10 + -0.26)
| Location | 9,917,528 – 9,917,648 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Shannon entropy | 0.36678 |
| G+C content | 0.47440 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -17.90 |
| Energy contribution | -17.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
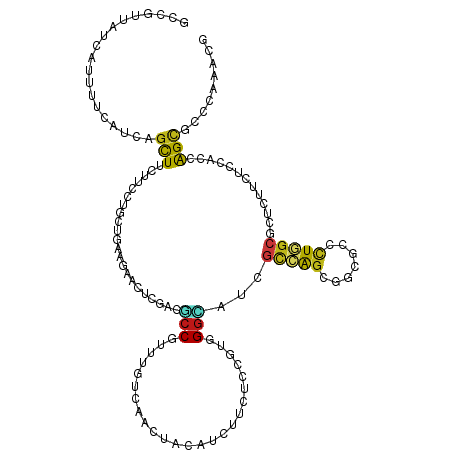
>dm3.chr2L 9917528 120 - 23011544 GCAUCCUGUGGAAGAAGAACGAGGUAGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUCAUUUUCAUCAGCUUCUUUAUCCUGAAGAACU ((....(((((((((((((.(.(((.(((........))).))).).))))...)))))))))..(((((((......))).)))).............))((((((.....)))))).. ( -33.40, z-score = -3.45, R) >droSim1.chr2L 9707097 120 - 22036055 GCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUUUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUCAUUUUCAUCAGCUUCUUCAUCCUGAAGAACU ((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............))((((((.....)))))).. ( -39.70, z-score = -4.93, R) >droSec1.super_3 5365073 120 - 7220098 GCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUUAUUUUCAUUAGCUUCUUCAUCCUGAAGAACU ((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............))((((((.....)))))).. ( -42.30, z-score = -5.90, R) >droYak2.chr2L 12608753 120 - 22324452 GCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUUAUUUUCAUCAGCUUCUUCAUCCUGAAGAACU ((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............))((((((.....)))))).. ( -42.30, z-score = -5.77, R) >droEre2.scaffold_4929 10533551 120 - 26641161 GCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUUAUUUUCAUCAGCUUCUUCAUCCUGAAGAACU ((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............))((((((.....)))))).. ( -42.30, z-score = -5.77, R) >droAna3.scaffold_12916 13757268 120 - 16180835 GUAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAAUAACGCCAUCUACCUGGCCGUGAUUAUCUUCAUCAGCUUCUUCCUGCUGAAGAACU ......(((((((((((((.(.(((((((........))))))).).))))...)))))))))...((((((......))).))).......(((.(((((........))))).))).. ( -43.20, z-score = -5.07, R) >dp4.chr4_group3 11438559 120 + 11692001 GCAUCCUCUGGAAGAAGAACGAGGUCGCCGACUACGAGGCCACCACCUUUUCGAUUUUCUACAACAAUUCCAUCUACCUGGCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAACU ......(((.(((((.(((((.(((.(((........))).))).........................(((......))).)))..)).))))).(((((........))))))))... ( -26.60, z-score = -0.96, R) >droPer1.super_1 8554696 120 + 10282868 GCAUCCUCUGGAAGAAGAACGAGGUCGCCGACUACGAGGCCACCACCUUUUCGAUUUUCUACAACAAUUCCAUCUACCUGGCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAACU ......(((.(((((.(((((.(((.(((........))).))).........................(((......))).)))..)).))))).(((((........))))))))... ( -26.60, z-score = -0.96, R) >droWil1.scaffold_180764 2526094 120 - 3949147 GCAUCUUGUGGAAAAAGAACGAAGUGGCUGACUAUGAAGCCACCACGUAUUCGAUUUUCUAUAACAAUGCCCUCUUCCUGGCUAUCAUCAUUUUCAUUAGCUUCUUUUUGCUAAAGAAUA ((((.((((((((((.(((((..((((((........))))))..)))..))..))))))))))..))))((.......))...........(((.(((((........))))).))).. ( -30.90, z-score = -3.14, R) >droVir3.scaffold_12963 14555001 120 - 20206255 GCAUCCUGUGGAAGAAGAACGAGGUUGCCGACUAUGAAGCGACCACAUACUCGAUUUUCUAUAACAACGCCCUCUACCUGGCCAUCAUCAUUUUCAUCAGCUUCUUCCUGCUGAAGAAUG ......(((...((((((.(((((((((..........))))))......))).))))))...)))..(((........)))..........(((.(((((........))))).))).. ( -26.50, z-score = -1.07, R) >droMoj3.scaffold_6500 17474826 120 - 32352404 GCAUCCUGUGGAAGAAGAAUGAAGUUGCCGACUACGAGGCCACGACCUAUUCGAUUUUCUACAACAAUGCCAUCUACUUGGCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAAUG ......(((((((((.(((((..((.(((........))).))....)))))..))))))))).....((((......))))..........(((.(((((........))))).))).. ( -32.50, z-score = -2.51, R) >droGri2.scaffold_15252 8251342 120 - 17193109 GCAUCCUGUGGAAGAAGAACGAGGUGGCUGACUAUGAGGCAACCACAUAUUCGAUUUUCUACAACAAUGCCAUCUAUUUGGCCAUCAUCAUCUUCAUCAGUUUCUUUCUGCUGAAGAACG ......(((((((((.(((....((((.((.((....)))).))))...)))..))))))))).....((((......))))........((((((.(((.......))).))))))... ( -31.90, z-score = -1.77, R) >anoGam1.chr3R 35788457 120 + 53272125 GGGUGCUGUGGAAGAAGAUCGACGUCGCCGAGUACGAGGCGACCACCUUCUCCAUCUUCUACAACAACGCCCUCUUCCUGGCCGUGGUCAUCUUUGCCAGCUUCUACCUGCUGCGAUCGU (((((.((((((((((((.....((((((........)))))).....)))...)))))))))....)))))......((((....))))...((((.(((........))))))).... ( -41.20, z-score = -1.78, R) >triCas2.chrUn_176 67431 120 + 68640 GGAUUUUGUGGAAGAAGAACGAAGUCGCGGAUUUUGAAGCCACAACUCUUUCAAUUUUCUACAACAACGCCUUGUUCUUGACUAUUGUUAUCAUUUCGAGCUUUUAUCUGCUCCCUUCGU .((((((((.........))))))))((((((...((((((..........((((..((...(((((....)))))...))..))))..........).))))).))))))......... ( -24.35, z-score = -0.28, R) >consensus GCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAACU ......(((((((((.((.....((.(((........))).)).......))..))))))))).....(((........)))..........(((.((((..........)))).))).. (-17.90 = -17.90 + -0.00)

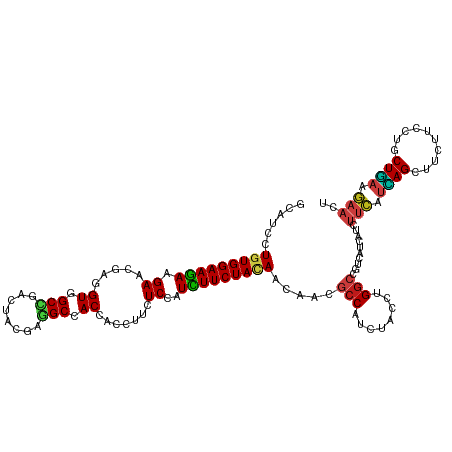
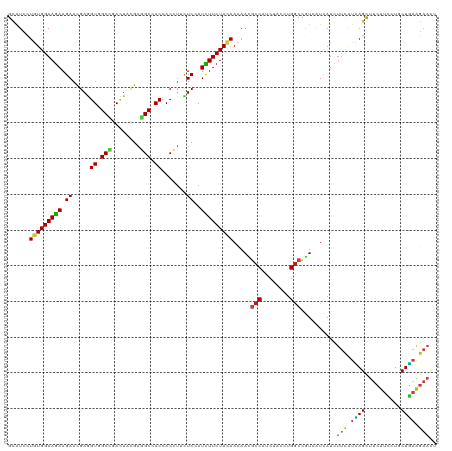
| Location | 9,917,530 – 9,917,650 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.25 |
| Shannon entropy | 0.35413 |
| G+C content | 0.48036 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.09 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9917530 120 - 23011544 GCGCAUCCUGUGGAAGAAGAACGAGGUAGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUCAUUUUCAUCAGCUUCUUUAUCCUGAAGAA ..((....(((((((((((((.(.(((.(((........))).))).).))))...)))))))))..(((((((......))).)))).............)).(((((.....))))). ( -33.20, z-score = -2.96, R) >droSim1.chr2L 9707099 120 - 22036055 GCGCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUUUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUCAUUUUCAUCAGCUUCUUCAUCCUGAAGAA ..((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............)).(((((.....))))). ( -39.50, z-score = -4.40, R) >droSec1.super_3 5365075 120 - 7220098 GCGCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUUAUUUUCAUUAGCUUCUUCAUCCUGAAGAA ..((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............)).(((((.....))))). ( -42.10, z-score = -5.38, R) >droYak2.chr2L 12608755 120 - 22324452 GCGCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUUAUUUUCAUCAGCUUCUUCAUCCUGAAGAA ..((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............)).(((((.....))))). ( -42.10, z-score = -5.23, R) >droEre2.scaffold_4929 10533553 120 - 26641161 GCGCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUUAUUUUCAUCAGCUUCUUCAUCCUGAAGAA ..((....(((((((((((((.(.(((((((........))))))).).))))...)))))))))..(((((((......))).)))).............)).(((((.....))))). ( -42.10, z-score = -5.23, R) >droAna3.scaffold_12916 13757270 120 - 16180835 GCGUAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAAUAACGCCAUCUACCUGGCCGUGAUUAUCUUCAUCAGCUUCUUCCUGCUGAAGAA (((.....(((((((((((((.(.(((((((........))))))).).))))...))))))))).....((((......))))))).....((((((.(((.......))).)))))). ( -44.10, z-score = -4.89, R) >dp4.chr4_group3 11438561 120 + 11692001 GCGCAUCCUCUGGAAGAAGAACGAGGUCGCCGACUACGAGGCCACCACCUUUUCGAUUUUCUACAACAAUUCCAUCUACCUGGCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAA ........(((.(((((.(((((.(((.(((........))).))).........................(((......))).)))..)).))))).(((((........)))))))). ( -26.60, z-score = -0.47, R) >droPer1.super_1 8554698 120 + 10282868 GCGCAUCCUCUGGAAGAAGAACGAGGUCGCCGACUACGAGGCCACCACCUUUUCGAUUUUCUACAACAAUUCCAUCUACCUGGCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAA ........(((.(((((.(((((.(((.(((........))).))).........................(((......))).)))..)).))))).(((((........)))))))). ( -26.60, z-score = -0.47, R) >droWil1.scaffold_180764 2526096 120 - 3949147 ACGCAUCUUGUGGAAAAAGAACGAAGUGGCUGACUAUGAAGCCACCACGUAUUCGAUUUUCUAUAACAAUGCCCUCUUCCUGGCUAUCAUCAUUUUCAUUAGCUUCUUUUUGCUAAAGAA ..((((.((((((((((.(((((..((((((........))))))..)))..))..))))))))))..))))((.......))...........(((.(((((........))))).))) ( -30.60, z-score = -2.96, R) >droVir3.scaffold_12963 14555003 120 - 20206255 ACGCAUCCUGUGGAAGAAGAACGAGGUUGCCGACUAUGAAGCGACCACAUACUCGAUUUUCUAUAACAACGCCCUCUACCUGGCCAUCAUCAUUUUCAUCAGCUUCUUCCUGCUGAAGAA .(((.....)))((((((((.(((((((((..........))))))......))).))))))........(((........)))..))......(((.(((((........))))).))) ( -26.00, z-score = -0.86, R) >droMoj3.scaffold_6500 17474828 120 - 32352404 ACGCAUCCUGUGGAAGAAGAAUGAAGUUGCCGACUACGAGGCCACGACCUAUUCGAUUUUCUACAACAAUGCCAUCUACUUGGCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAA ........(((((((((.(((((..((.(((........))).))....)))))..))))))))).....((((......))))........((((((.(((.......))).)))))). ( -32.10, z-score = -2.37, R) >droGri2.scaffold_15252 8251344 120 - 17193109 ACGCAUCCUGUGGAAGAAGAACGAGGUGGCUGACUAUGAGGCAACCACAUAUUCGAUUUUCUACAACAAUGCCAUCUAUUUGGCCAUCAUCAUCUUCAUCAGUUUCUUUCUGCUGAAGAA ........(((((((((.(((....((((.((.((....)))).))))...)))..))))))))).....((((......))))........((((((.(((.......))).)))))). ( -31.90, z-score = -1.63, R) >anoGam1.chr3R 35788459 120 + 53272125 GCGGGUGCUGUGGAAGAAGAUCGACGUCGCCGAGUACGAGGCGACCACCUUCUCCAUCUUCUACAACAACGCCCUCUUCCUGGCCGUGGUCAUCUUUGCCAGCUUCUACCUGCUGCGAUC ..(((((.((((((((((((.....((((((........)))))).....)))...)))))))))....)))))......((((....))))...((((.(((........))))))).. ( -41.50, z-score = -1.88, R) >triCas2.chrUn_176 67433 120 + 68640 GCGGAUUUUGUGGAAGAAGAACGAAGUCGCGGAUUUUGAAGCCACAACUCUUUCAAUUUUCUACAACAACGCCUUGUUCUUGACUAUUGUUAUCAUUUCGAGCUUUUAUCUGCUCCCUUC ...((((((((.........))))))))((((((...((((((..........((((..((...(((((....)))))...))..))))..........).))))).))))))....... ( -24.35, z-score = -0.32, R) >consensus GCGCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAA ..((....(((((((((.((.....((.(((........))).)).......))..))))))))).....(((........))).................)).(((((.....))))). (-17.85 = -18.09 + 0.24)
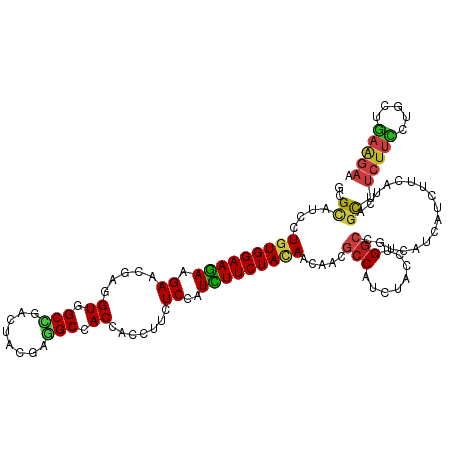
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:16 2011