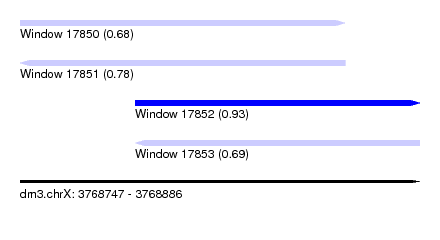
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,768,747 – 3,768,886 |
| Length | 139 |
| Max. P | 0.933681 |

| Location | 3,768,747 – 3,768,860 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.50733 |
| G+C content | 0.41399 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -11.99 |
| Energy contribution | -15.31 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
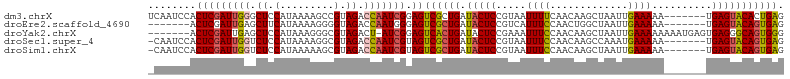
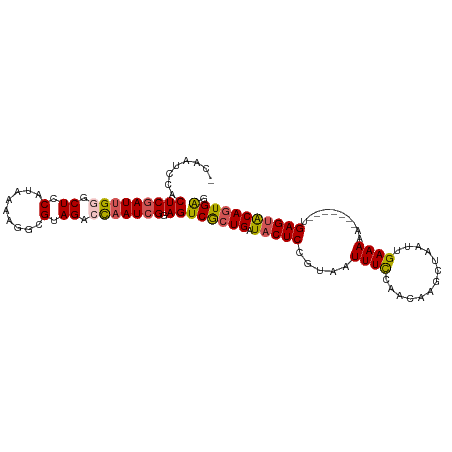
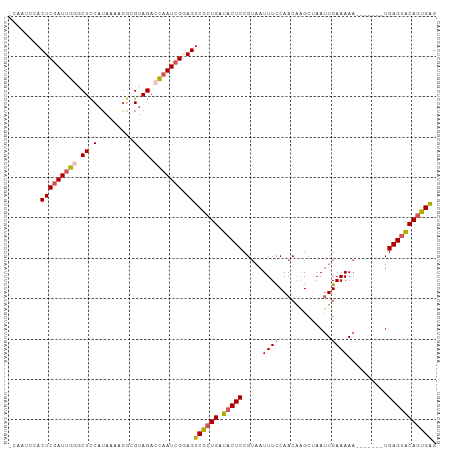
>dm3.chrX 3768747 113 + 22422827 AGGGCAUACCAGGCUUAUCUAAUUAUUUAUAGGUAAAGCCCUCAGUGUACUCA-------UUUUUCAAUUAGCUUGUUGAAAAUUACGGAGUAUCAGCGACUCCGAUUGGUCUACGGCUU .((((......((((((((((........))))).)))))....(((.(((..-------.((((((((......))))))))...((((((.......))))))...))).))).)))) ( -28.90, z-score = -1.55, R) >droEre2.scaffold_4690 1152798 113 + 18748788 AAGGCAUGCCAGGCUUAUCUAAUUAUUUAUAGGUAAAGCCCUCACUGUACUCA-------UUUUUCAAUUAGCCAGUUGGAAAUGACGGAGUAUCAGCGACUCCCAUUGGUCUACCCCUU ((((...((((((((((((((........))))).))))).((.((((((((.-------.(((((((((....))))))))).....))))).))).)).......)))).....)))) ( -30.00, z-score = -1.78, R) >droYak2.chrX 17124524 119 - 21770863 AAGGCAUACCAGGCUUAUUUAAUUAUUUAUAGGCAAAGCCCCCACUGCCCUCACUCAUUUUUUUUCAAUUAGCUUGUUGGAAAUUUCGGAGUAUCAGUGACUCCGAU-AGUCUACGCCCU ..(((....((((((.((((((....)))..((((..........)))).................))).)))))).((((....(((((((.......))))))).-..)))).))).. ( -26.50, z-score = -1.35, R) >droSec1.super_4 3240516 113 - 6179234 AAGGCAUACCAGGCUUAUCUAAUUAUUUAUAGGCAAAGCCCUCACUGUACUCA-------UUUUUCAUUUGGCUUGUUGGAAAUUACGGAGUAUCAGCGACUACGAUUGGUCUACGCCUU (((((..(((((((((.((((........))))..))))).((.((((((((.-------.((((((..........)))))).....))))).))).)).......))))....))))) ( -27.90, z-score = -1.53, R) >droSim1.chrX 2774721 113 + 17042790 AAGGCAUACCAGGCUUAUCUAAUUAUUUAUAGGUAAAGCCCUCACUGUACUCA-------UUUUUCAAUUAGCUUGUUGGAAAUUACGGAGUAUCAGCGACUACGAUUGGUCUACGCUUU (((((..((((((((((((((........))))).))))).((.((((((((.-------.((((((((......)))))))).....))))).))).)).......))))....))))) ( -30.80, z-score = -2.89, R) >droPer1.super_25 250627 91 + 1448063 GAUCCGCAGUAGAGUGACCU------UCAAAAAUGGUGACCCCAUGCCGCUUG-------------AAUUUG---AAUGCAAUUUGAAGAGCAUUA-CAGCUCCGA---GUCCAUCU--- ((..((.(((.(((((..((------(((((...(((........)))((...-------------......---...))..)))))))..)))).-).))).)).---.)).....--- ( -20.10, z-score = -1.09, R) >consensus AAGGCAUACCAGGCUUAUCUAAUUAUUUAUAGGUAAAGCCCUCACUGUACUCA_______UUUUUCAAUUAGCUUGUUGGAAAUUACGGAGUAUCAGCGACUCCGAUUGGUCUACGCCUU .((((......((((((((((........))))))).))).((.((((((((.........((((((((......)))))))).....))))).))).)).........))))....... (-11.99 = -15.31 + 3.31)

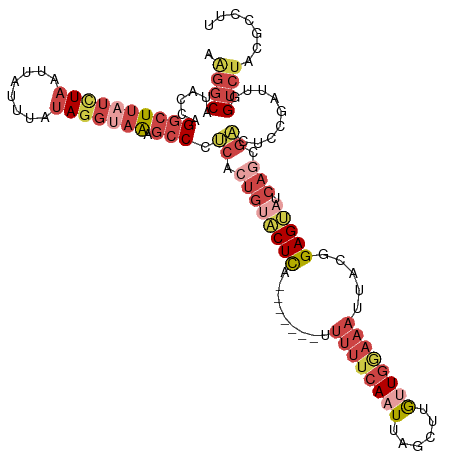
| Location | 3,768,747 – 3,768,860 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.99 |
| Shannon entropy | 0.50733 |
| G+C content | 0.41399 |
| Mean single sequence MFE | -27.95 |
| Consensus MFE | -14.88 |
| Energy contribution | -15.97 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.779832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3768747 113 - 22422827 AAGCCGUAGACCAAUCGGAGUCGCUGAUACUCCGUAAUUUUCAACAAGCUAAUUGAAAAA-------UGAGUACACUGAGGGCUUUACCUAUAAAUAAUUAGAUAAGCCUGGUAUGCCCU .....(((.((((..((((((.......)))))).....((((....(((.(((....))-------).)))....))))(((((...(((........)))..))))))))).)))... ( -24.70, z-score = -1.04, R) >droEre2.scaffold_4690 1152798 113 - 18748788 AAGGGGUAGACCAAUGGGAGUCGCUGAUACUCCGUCAUUUCCAACUGGCUAAUUGAAAAA-------UGAGUACAGUGAGGGCUUUACCUAUAAAUAAUUAGAUAAGCCUGGCAUGCCUU ..((((((..((........((((((.(((((.((((........))))..(((....))-------))))))))))))((((((...(((........)))..))))))))..)))))) ( -33.60, z-score = -1.84, R) >droYak2.chrX 17124524 119 + 21770863 AGGGCGUAGACU-AUCGGAGUCACUGAUACUCCGAAAUUUCCAACAAGCUAAUUGAAAAAAAAUGAGUGAGGGCAGUGGGGGCUUUGCCUAUAAAUAAUUAAAUAAGCCUGGUAUGCCUU ((((((((....-.(((((((.......))))))).........((.(((..((....))..........((((((........))))))...............))).)).)))))))) ( -28.90, z-score = -0.44, R) >droSec1.super_4 3240516 113 + 6179234 AAGGCGUAGACCAAUCGUAGUCGCUGAUACUCCGUAAUUUCCAACAAGCCAAAUGAAAAA-------UGAGUACAGUGAGGGCUUUGCCUAUAAAUAAUUAGAUAAGCCUGGUAUGCCUU (((((((..(((........((((((.(((((.....((((.............))))..-------.)))))))))))((((((...(((........)))..)))))))))))))))) ( -31.32, z-score = -2.80, R) >droSim1.chrX 2774721 113 - 17042790 AAAGCGUAGACCAAUCGUAGUCGCUGAUACUCCGUAAUUUCCAACAAGCUAAUUGAAAAA-------UGAGUACAGUGAGGGCUUUACCUAUAAAUAAUUAGAUAAGCCUGGUAUGCCUU .(((.(((.(((........((((((.(((((.((........))......(((....))-------))))))))))))((((((...(((........)))..))))))))).)))))) ( -28.10, z-score = -2.41, R) >droPer1.super_25 250627 91 - 1448063 ---AGAUGGAC---UCGGAGCUG-UAAUGCUCUUCAAAUUGCAUU---CAAAUU-------------CAAGCGGCAUGGGGUCACCAUUUUUGA------AGGUCACUCUACUGCGGAUC ---.....((.---.((((..((-.(((((..........)))))---))..))-------------)..((((..((((((.(((........------.))).)))))))))))..)) ( -21.10, z-score = -0.01, R) >consensus AAGGCGUAGACCAAUCGGAGUCGCUGAUACUCCGUAAUUUCCAACAAGCUAAUUGAAAAA_______UGAGUACAGUGAGGGCUUUACCUAUAAAUAAUUAGAUAAGCCUGGUAUGCCUU .....(((.(((........((((((.(((((....................................)))))))))))((((((...(((........)))..))))))))).)))... (-14.88 = -15.97 + 1.09)

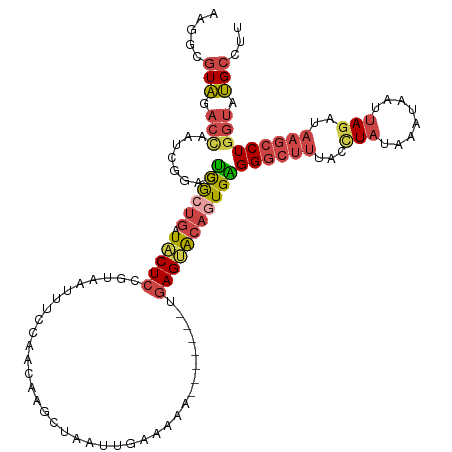
| Location | 3,768,787 – 3,768,886 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.50 |
| Shannon entropy | 0.27314 |
| G+C content | 0.42691 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -17.88 |
| Energy contribution | -19.08 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.933681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3768787 99 + 22422827 CUCAGUGUACUCA-------UUUUUCAAUUAGCUUGUUGAAAAUUACGGAGUAUCAGCGACUCCGAUUGGUCUACGGCUUUUAUGGAGCCCAAUCGAGUGGAUUGA .(((((.(((((.-------.((((((((......))))))))....(((((.......)))))((((((....)(((((.....))))))))))))))).))))) ( -32.80, z-score = -3.37, R) >droEre2.scaffold_4690 1152838 92 + 18748788 CUCACUGUACUCA-------UUUUUCAAUUAGCCAGUUGGAAAUGACGGAGUAUCAGCGACUCCCAUUGGUCUACCCCUUUUAUGAAGCUCAAUCGAGU------- .(((((((((((.-------.(((((((((....))))))))).....))))).))).((((......))))...........))).((((....))))------- ( -19.30, z-score = -0.59, R) >droYak2.chrX 17124564 98 - 21770863 CCCACUGCCCUCACUCAUUUUUUUUCAAUUAGCUUGUUGGAAAUUUCGGAGUAUCAGUGACUCCGAU-AGUCUACGCCCUUUAUGGAGCUCAAUCGAGU------- ..(((((.....((((.....((((((((......)))))))).....))))..)))))((((.(((-.(.....((((.....)).)).).)))))))------- ( -22.10, z-score = -1.18, R) >droSec1.super_4 3240556 98 - 6179234 CUCACUGUACUCA-------UUUUUCAUUUGGCUUGUUGGAAAUUACGGAGUAUCAGCGACUACGAUUGGUCUACGCCUUUUAUGGAGACCAAUCGAGUGGAUUG- ....((((((((.-------.((((((..........)))))).....))))).)))(.(((.((((((((((...((......))))))))))))))).)....- ( -30.50, z-score = -3.31, R) >droSim1.chrX 2774761 98 + 17042790 CUCACUGUACUCA-------UUUUUCAAUUAGCUUGUUGGAAAUUACGGAGUAUCAGCGACUACGAUUGGUCUACGCUUUUUAUGGAGACCAAUCGAGUGGAUUG- ....((((((((.-------.((((((((......)))))))).....))))).)))(.(((.((((((((((.((.......)).))))))))))))).)....- ( -31.90, z-score = -4.20, R) >consensus CUCACUGUACUCA_______UUUUUCAAUUAGCUUGUUGGAAAUUACGGAGUAUCAGCGACUCCGAUUGGUCUACGCCUUUUAUGGAGCCCAAUCGAGUGGAUUG_ ..(.((((((((.........((((((((......)))))))).....))))).))).)(((.(((((((.((.((.......)).)).))))))))))....... (-17.88 = -19.08 + 1.20)
| Location | 3,768,787 – 3,768,886 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Shannon entropy | 0.27314 |
| G+C content | 0.42691 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3768787 99 - 22422827 UCAAUCCACUCGAUUGGGCUCCAUAAAAGCCGUAGACCAAUCGGAGUCGCUGAUACUCCGUAAUUUUCAACAAGCUAAUUGAAAAA-------UGAGUACACUGAG (((....(((((((((((((.......))))(....))))))(((((.......)))))....(((((((........))))))).-------.))))....))). ( -21.50, z-score = -1.44, R) >droEre2.scaffold_4690 1152838 92 - 18748788 -------ACUCGAUUGAGCUUCAUAAAAGGGGUAGACCAAUGGGAGUCGCUGAUACUCCGUCAUUUCCAACUGGCUAAUUGAAAAA-------UGAGUACAGUGAG -------.(((.((((.(((((......)))))....)))))))..((((((.(((((.((((........))))..(((....))-------)))))))))))). ( -22.00, z-score = -0.60, R) >droYak2.chrX 17124564 98 + 21770863 -------ACUCGAUUGAGCUCCAUAAAGGGCGUAGACU-AUCGGAGUCACUGAUACUCCGAAAUUUCCAACAAGCUAAUUGAAAAAAAAUGAGUGAGGGCAGUGGG -------...(.((((.((((......)))).......-.(((((((.......))))))).....((.....(((.(((.......))).)))...)))))).). ( -19.20, z-score = 0.55, R) >droSec1.super_4 3240556 98 + 6179234 -CAAUCCACUCGAUUGGUCUCCAUAAAAGGCGUAGACCAAUCGUAGUCGCUGAUACUCCGUAAUUUCCAACAAGCCAAAUGAAAAA-------UGAGUACAGUGAG -.......((((((((((((((......))...)))))))))).))((((((.(((((.....((((.............))))..-------.))))))))))). ( -28.02, z-score = -4.40, R) >droSim1.chrX 2774761 98 - 17042790 -CAAUCCACUCGAUUGGUCUCCAUAAAAAGCGUAGACCAAUCGUAGUCGCUGAUACUCCGUAAUUUCCAACAAGCUAAUUGAAAAA-------UGAGUACAGUGAG -.......((((((((((((.(.........).)))))))))).))((((((.(((((.((........))......(((....))-------)))))))))))). ( -27.20, z-score = -4.55, R) >consensus _CAAUCCACUCGAUUGGGCUCCAUAAAAGGCGUAGACCAAUCGGAGUCGCUGAUACUCCGUAAUUUCCAACAAGCUAAUUGAAAAA_______UGAGUACAGUGAG ........(((((((((.((.(.........).)).))))))).))((((((.(((((............(((.....))).............))))))))))). (-14.89 = -15.73 + 0.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:32 2011