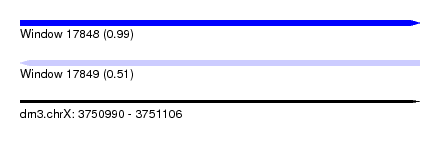
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,750,990 – 3,751,106 |
| Length | 116 |
| Max. P | 0.993637 |

| Location | 3,750,990 – 3,751,106 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Shannon entropy | 0.32798 |
| G+C content | 0.49201 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -24.54 |
| Energy contribution | -26.04 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
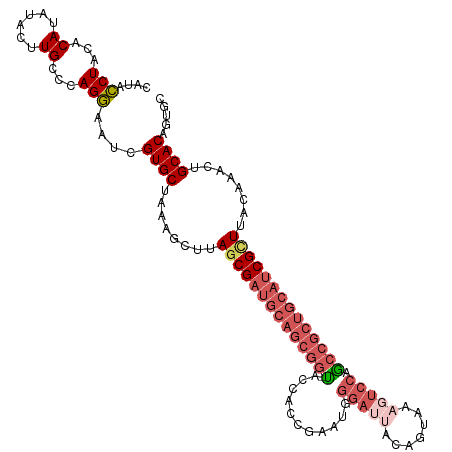
>dm3.chrX 3750990 116 + 22422827 CAUACCUACACAUAUACUUGCCCAGGAAUCGUGCUAAAGCUUAGCGAUGCAGCGG-CACCACCGAAUGGGAUUGCAGUGAAGUCCAGCCGCUGCAACGCUUACGAACUGCACAGUGC ........(((.....(((....)))....((((........((((.((((((((-(..((.....))(((((.......))))).))))))))).))))........)))).))). ( -39.19, z-score = -2.08, R) >droEre2.scaffold_4690 1134140 94 + 18748788 CAUACCUACACAUAUACUUGCCCAGGCAUUGUGCUAAUUCUUAGCGAG--------UACGA---------------GUAAAAUCCAACCGCUACAUCGUUUACAAGCUGCACAGUGC .........................(((((((((........((((((--------((((.---------------((........)))).))).)))))........))))))))) ( -22.09, z-score = -1.66, R) >droYak2.chrX 17104522 117 - 21770863 CAUAUCUACACAUAUACUUGCCCAGAAAUUGUGCUUAUUCUUAGCGAUGCAGCGGUUACCACCGAUUGGGACCAUAGUAAAAUCCCACCGCUGCAUCGUUUACCAACUGCACAGUGC ....(((...((......))...))).(((((((........((((((((((((((...........((((...........))))))))))))))))))........))))))).. ( -37.19, z-score = -4.40, R) >droSec1.super_4 3216674 116 - 6179234 CAUACCUACACAUAUACUUGCCCAGGAAUCGUGCUAAAGCUUACCGAUGCAGCGG-UACCACCGAAUGGGAUUACAGUAUAGUCCAGCCGCUGCAUCGCUUACAAACUGCACAGUGC ........(((.....(((....)))....((((..........(((((((((((-(..((.....))((((((.....)))))).))))))))))))..........)))).))). ( -36.05, z-score = -2.50, R) >droSim1.chrX 9976866 116 + 17042790 CAUACCUACACAUAUACUUGCCCAGGAAUCGUGCGAAAGCUUAGCGAUGCAGCGG-UACCACCGAAUGGGAUUACAGUAUAGUCCAGCCGCUGCAUCGCUUACAAGCUGCACAGUGC ........(((.....(((....)))....((((...((((((((((((((((((-(..((.....))((((((.....)))))).))))))))))))))...))))))))).))). ( -44.70, z-score = -4.12, R) >consensus CAUACCUACACAUAUACUUGCCCAGGAAUCGUGCUAAAGCUUAGCGAUGCAGCGG_UACCACCGAAUGGGAUUACAGUAAAGUCCAGCCGCUGCAUCGCUUACAAACUGCACAGUGC ....(((...((......))...)))....((((........(((((((((((((...........((......))...........)))))))))))))........))))..... (-24.54 = -26.04 + 1.50)

| Location | 3,750,990 – 3,751,106 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Shannon entropy | 0.32798 |
| G+C content | 0.49201 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -26.71 |
| Energy contribution | -28.29 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.507466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
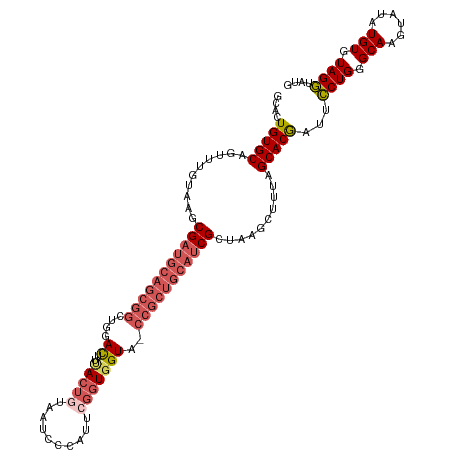
>dm3.chrX 3750990 116 - 22422827 GCACUGUGCAGUUCGUAAGCGUUGCAGCGGCUGGACUUCACUGCAAUCCCAUUCGGUGGUG-CCGCUGCAUCGCUAAGCUUUAGCACGAUUCCUGGGCAAGUAUAUGUGUAGGUAUG (((((((.(((.((((.((((.(((((((((......((((((.(((...))))))))).)-)))))))).))))..((....))))))...))).))).......))))....... ( -42.61, z-score = -1.02, R) >droEre2.scaffold_4690 1134140 94 - 18748788 GCACUGUGCAGCUUGUAAACGAUGUAGCGGUUGGAUUUUAC---------------UCGUA--------CUCGCUAAGAAUUAGCACAAUGCCUGGGCAAGUAUAUGUGUAGGUAUG ....(((((...(((....)))..(((((((..((......---------------))..)--------).))))).......)))))(((((((.(((......))).))))))). ( -24.90, z-score = 0.08, R) >droYak2.chrX 17104522 117 + 21770863 GCACUGUGCAGUUGGUAAACGAUGCAGCGGUGGGAUUUUACUAUGGUCCCAAUCGGUGGUAACCGCUGCAUCGCUAAGAAUAAGCACAAUUUCUGGGCAAGUAUAUGUGUAGAUAUG (((((((((.(((....)))(((((((((((((((((.......)))))).)))(((....)))))))))))(((.((((..........)))).)))..))))).))))....... ( -43.00, z-score = -3.28, R) >droSec1.super_4 3216674 116 + 6179234 GCACUGUGCAGUUUGUAAGCGAUGCAGCGGCUGGACUAUACUGUAAUCCCAUUCGGUGGUA-CCGCUGCAUCGGUAAGCUUUAGCACGAUUCCUGGGCAAGUAUAUGUGUAGGUAUG ....(((((((((((....(((((((((((....(((((...((......))...))))).-))))))))))).))))))...)))))...((((.(((......))).)))).... ( -39.10, z-score = -0.65, R) >droSim1.chrX 9976866 116 - 17042790 GCACUGUGCAGCUUGUAAGCGAUGCAGCGGCUGGACUAUACUGUAAUCCCAUUCGGUGGUA-CCGCUGCAUCGCUAAGCUUUCGCACGAUUCCUGGGCAAGUAUAUGUGUAGGUAUG ....((((((((((...(((((((((((((....(((((...((......))...))))).-))))))))))))))))))...)))))...((((.(((......))).)))).... ( -45.80, z-score = -2.36, R) >consensus GCACUGUGCAGUUUGUAAGCGAUGCAGCGGCUGGACUUUACUGUAAUCCCAUUCGGUGGUA_CCGCUGCAUCGCUAAGCUUUAGCACGAUUCCUGGGCAAGUAUAUGUGUAGGUAUG ....(((((..........(((((((((((....((..(((((..........)))))))..)))))))))))..........)))))...((((.(((......))).)))).... (-26.71 = -28.29 + 1.58)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:29 2011