
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,743,839 – 3,743,947 |
| Length | 108 |
| Max. P | 0.796037 |

| Location | 3,743,839 – 3,743,947 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 64.79 |
| Shannon entropy | 0.68734 |
| G+C content | 0.48988 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -7.78 |
| Energy contribution | -8.55 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3743839 108 - 22422827 AACCAUCAUUUGGCACACC------CUAA-------GCCUUA-AACGC-----AACUCGACUGGACUUGACUUUCGCUUUUGGCCUGCUCUCGACCUCAUUUGCGG-CCAAUCGAAAGUUUCGCCAGC ...........(((.....------....-------)))...-.....-----.......((((.(..((((((((...((((((.((..............))))-)))).))))))))..))))). ( -29.44, z-score = -2.67, R) >droPer1.super_25 218194 94 - 1448063 ---AACAGCAACAACUGGCCGAAGA-------------------ACAG-----AGCUUGGCU-UUCGGGACUUUCGCUUUUGG-----UCUCGACCUCAUUUGCGGGCCGAUCGAAAGUUUCGUUUC- ---..(((......)))(((((...-------------------....-----...))))).-..((..(((((((...((((-----((((((......))).))))))).)))))))..))....- ( -28.40, z-score = -0.78, R) >droAna3.scaffold_12929 306397 89 - 3277472 --GUUCGUCAAUGCACACCCAUAUA------------------UAUAC-----AGACUGACU------GACUUUCGC-UUUGG-----UCUCGACCUCA-UUAAGGCCCAAUCGAAAGUUUCGAUCU- --.......................------------------.....-----(((.(((..------((((((((.-((.((-----(((..(.....-)..))))).)).))))))))))).)))- ( -17.90, z-score = -1.33, R) >droEre2.scaffold_4690 1126962 116 - 18748788 AGCCAUCAUUUGGCACACCCUAAGCCUUA-------ACAGCGCAACUCG----AACUCGACUGGACUUGACUUUCGCUUUUGGCCCGCUCUCGACCUCAUUUGCGG-CCAAUCGAAAGUUUCGCCAGU .((((.....)))).........((((..-------..)).))...(((----....)))((((.(..((((((((...((((((.((..............))))-)))).))))))))..))))). ( -35.64, z-score = -2.92, R) >droYak2.chrX 17097348 127 + 21770863 AACCAUCAUUUGGCACACCCUAAGCCUUAAACACACACAGCAAAACACAACACAGUUCGACUGGACUUGACUUUCGCUUUUGGCCUGCUCUCGACCUCAUUUGCGG-CCAAUCGAAAGUUUCGCCAGU ...........(((.........))).................................(((((.(..((((((((...((((((.((..............))))-)))).))))))))..)))))) ( -30.64, z-score = -2.37, R) >droSec1.super_4 3209551 113 + 6179234 AACCAUCAUUUGGCACACCCUACC-CUAA-------GCCUUA-AACGC-----AACUCGACUGGACUUGACUUUCGCUUUUGGCCUGCUCUCGACCUCAUUUGCGG-CCAAUCGAAAGUUUCGCCAGC ...........(((..........-....-------)))...-.....-----.......((((.(..((((((((...((((((.((..............))))-)))).))))))))..))))). ( -28.98, z-score = -2.55, R) >droSim1.chrX 2749533 114 - 17042790 AACCAUCAUUUGGCACACCCUACCACUAA-------GCCUUA-AACGC-----AACUCGACUGGACUUGACUUUCGCUUUUGGCCUGCUCUCGACCUCAUUUGCGG-CCAAUCGAAAGUUUCGCCAGC ...........(((...............-------)))...-.....-----.......((((.(..((((((((...((((((.((..............))))-)))).))))))))..))))). ( -28.90, z-score = -2.64, R) >anoGam1.chr2L 22295101 104 + 48795086 --AAACCUUUUAGAAUAACCGAAUCCUU----------------UCUC-----ACUUCGAACGGAAUUACCCUGCGUGUUUGGUUAUUUUCUAAUCUCAUUCGCGG-CCCAUCGAAACUUGCCCAAGC --.......(((((((((((((((((.(----------------((..-----.....))).)))..(((.....))))))))))))..)))))........(.((-(............)))).... ( -16.40, z-score = -0.10, R) >consensus AACCAUCAUUUGGCACACCCUAAGACUAA________C___A_AACAC_____AACUCGACUGGACUUGACUUUCGCUUUUGGCCUGCUCUCGACCUCAUUUGCGG_CCAAUCGAAAGUUUCGCCAGC ....................................................................((((((((...((((((...................)).)))).))))))))........ ( -7.78 = -8.55 + 0.77)
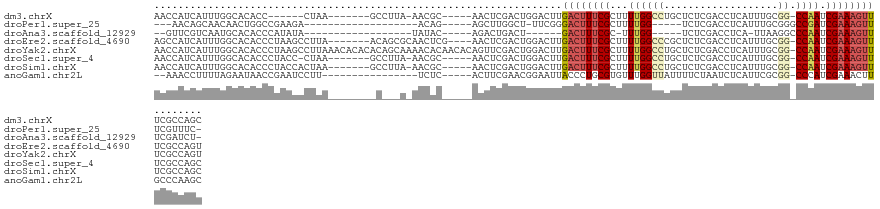

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:27 2011