
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,741,330 – 3,741,474 |
| Length | 144 |
| Max. P | 0.531913 |

| Location | 3,741,330 – 3,741,474 |
|---|---|
| Length | 144 |
| Sequences | 6 |
| Columns | 155 |
| Reading direction | reverse |
| Mean pairwise identity | 69.79 |
| Shannon entropy | 0.51519 |
| G+C content | 0.48513 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -16.75 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.531913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
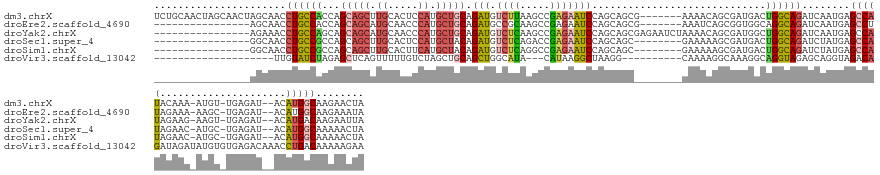
>dm3.chrX 3741330 144 - 22422827 UCUGCAACUAGCAACUAGCAACCUGCCACCAGCAGCUUGCACUCCAUGCUGCAGAUGUCUUAAGCCGAGAAUCCAGCAGCG-------AAAACAGCGAUGACUGGCAGAUCAAUGAGCCAUACAAA-AUGU-UGAGAU--ACAUGGCAAGAACUA ((((((....(((....((((.((((.....)))).))))......)))))))))..((((..((((.(.((((((((((.-------......))..((..((((..........))))..))..-.)))-)).)))--.).)))))))).... ( -38.20, z-score = -0.65, R) >droEre2.scaffold_4690 1124546 128 - 18748788 ----------------AGCAACCUGCCACCAGCAGCAUGCAACCCAUGCUGCAGAUGCCGCAAGCCGAGAAUCCAGCAGCG-------AAAUCAGCGGUGGCAGGCAGAUCAAUGAGCCUUAGAAA-AAGC-UGAGAU--ACAUGGCAAGAAAUA ----------------.((..(((((((((.((((((((.....)))))))).(((..(((..((.(......).)).)))-------..)))...))))))))).......(((...(((((...-...)-))))..--.))).))........ ( -47.00, z-score = -3.64, R) >droYak2.chrX 17094852 135 + 21770863 ----------------AGAAACCUGCCAGCAGCAGCAUGCAACCCAUGCUGCAGAUGUCUCAAGCCGAGAAUCCAGCAGCGAGAAUCUAAAACAGCGAUGGCUGGCAGAUCAAUGAGCCAUAGAAG-AAGU-UGAGAU--ACAUGACAAGAAUUA ----------------......((((((((.((((((((.....))))))))((((.((((..((.(......).))...))))))))............)))))))).(((((...(.......)-..))-)))...--............... ( -38.90, z-score = -2.35, R) >droSec1.super_4 3207070 127 + 6179234 ----------------GGCAACCUGCCGCCAGCAGCUUGCACUCCAUGCUACAGAUGUCUCAGACCGAGAAUCCAGCAGC--------GAAAAAGCGAUGACUGGCAGAUCUAUGAGCCAUAGAAC-AUGC-UGAGAU--ACAUGGCAAAAACUA ----------------.((((.((((.....)))).))))......(((((....((((((((.........((((((.(--------(......)).)).))))..(.((((((...)))))).)-...)-))))))--)..)))))....... ( -36.00, z-score = -1.20, R) >droSim1.chrX 2747031 127 - 17042790 ----------------GGCAACCUGCCGCCAGCAGCUUGCACUUCAUGCUACAGAUGUCUCAGGCCGAGAAUCCAGCAGC--------GAAAAAGCGAUGACUGGCAGAUCUAUGAGCCAUAGAAC-AUGC-UGAGAU--ACAUGGCAAAAACUA ----------------.((((.((((.....)))).))))......(((((....((((((((((((...(((.....((--------......)))))...)))).(.((((((...)))))).)-...)-))))))--)..)))))....... ( -36.50, z-score = -0.62, R) >droVir3.scaffold_13042 3431036 122 - 5191987 --------------------UUGUAUCUAGAGCUCAGUUUUUGUCUAGCUGCAGCUGGCAUA---CAUAAGGCUAAGG----------CAAAAGGCAAAGGCAGGUAGAGCAGGUAGACAGAUAGAUAUGUGUGAGACAAACCUGACAAAAAGAA --------------------..((((((((....)....((((((((.((((.((((((...---......)))..))----------).....((....)).......)))).))))))))))))))).(((.((......)).)))....... ( -31.00, z-score = -0.55, R) >consensus ________________AGCAACCUGCCACCAGCAGCUUGCACUCCAUGCUGCAGAUGUCUCAAGCCGAGAAUCCAGCAGC________AAAAAAGCGAUGACUGGCAGAUCAAUGAGCCAUAGAAA_AUGC_UGAGAU__ACAUGGCAAAAACUA ......................((((((...(((((.((.....)).))))).(((.((((.....))))))).............................))))))........(((((.....................)))))........ (-16.75 = -17.40 + 0.65)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:24 2011