
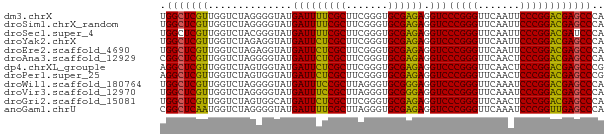
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,721,653 – 3,721,727 |
| Length | 74 |
| Max. P | 0.992808 |

| Location | 3,721,653 – 3,721,727 |
|---|---|
| Length | 74 |
| Sequences | 12 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 93.37 |
| Shannon entropy | 0.14660 |
| G+C content | 0.60360 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -30.72 |
| Energy contribution | -29.97 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
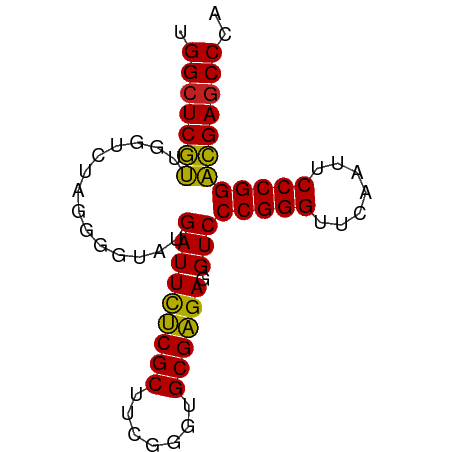
>dm3.chrX 3721653 74 + 22422827 UGGCUCGUUGGUCUAGGGGUAUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCA .((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).. ( -30.60, z-score = -1.41, R) >droSim1.chrX_random 1513385 74 + 5698898 UGGCUCGUUGGUCUAGGGGUAUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCA .((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).. ( -30.60, z-score = -1.41, R) >droSec1.super_4 3188024 74 - 6179234 UGGCUCGUUGGUCUACGGGUAUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAUCCCA ..((((((......)))))).....((((((.......))))))(((((((((.......)))))..))))... ( -24.80, z-score = 0.26, R) >droYak2.chrX 17075043 74 - 21770863 UGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCA .(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).. ( -31.50, z-score = -1.77, R) >droEre2.scaffold_4690 1104880 74 + 18748788 UGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCA .(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).. ( -31.50, z-score = -1.77, R) >droAna3.scaffold_12929 283475 74 + 3277472 CGGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCCA .((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).. ( -33.50, z-score = -1.60, R) >dp4.chrXL_group1e 6860292 74 + 12523060 AGGCUCGUUGGUCUAGUGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCCG .(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).. ( -31.70, z-score = -1.63, R) >droPer1.super_25 195702 74 + 1448063 AGGCUCGUUGGUCUAGUGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCCG .(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).. ( -31.70, z-score = -1.63, R) >droWil1.scaffold_180764 989789 74 - 3949147 UGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCA .((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).. ( -32.40, z-score = -1.53, R) >droVir3.scaffold_12970 6178752 74 - 11907090 UGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCA .((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).. ( -32.40, z-score = -1.53, R) >droGri2.scaffold_15081 1374012 74 - 4274704 UGGCUCGUUGGUCUAGUGGCAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAACUCCCGGACGAGCCCA .(((((((..(((....)))..(((((((((.......)))))).)))(((((.......)))))))))))).. ( -33.20, z-score = -1.94, R) >anoGam1.chrU 1854101 74 + 59568033 CGGCUCAAUGGUCUAGGGGUAUGAUUUUCGCUUAGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCA .((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).. ( -29.40, z-score = -1.82, R) >consensus UGGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCA .(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).. (-30.72 = -29.97 + -0.75)
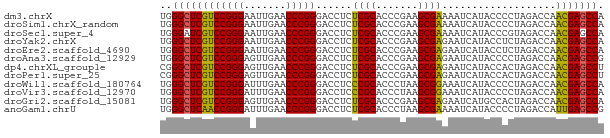
| Location | 3,721,653 – 3,721,727 |
|---|---|
| Length | 74 |
| Sequences | 12 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 93.37 |
| Shannon entropy | 0.14660 |
| G+C content | 0.60360 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -26.11 |
| Energy contribution | -25.61 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.992808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3721653 74 - 22422827 UGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAUACCCCUAGACCAACGAGCCA ..((((((((((((.......)))))......((((.......))))...................))))))). ( -25.90, z-score = -2.37, R) >droSim1.chrX_random 1513385 74 - 5698898 UGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAUACCCCUAGACCAACGAGCCA ..((((((((((((.......)))))......((((.......))))...................))))))). ( -25.90, z-score = -2.37, R) >droSec1.super_4 3188024 74 + 6179234 UGGGAUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAUACCCGUAGACCAACGAGCCA ..((.(((((((((.......)))))......((((.......))))...................)))).)). ( -19.30, z-score = 0.21, R) >droYak2.chrX 17075043 74 + 21770863 UGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCA ..((((((((((((.......)))))((..((((((.......)))))).))..............))))))). ( -30.90, z-score = -3.62, R) >droEre2.scaffold_4690 1104880 74 - 18748788 UGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCA ..((((((((((((.......)))))((..((((((.......)))))).))..............))))))). ( -30.90, z-score = -3.62, R) >droAna3.scaffold_12929 283475 74 - 3277472 UGGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCG ..((((((((((((.......)))))((..((((((.......)))))).))..............))))))). ( -30.90, z-score = -3.13, R) >dp4.chrXL_group1e 6860292 74 - 12523060 CGGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCACUAGACCAACGAGCCU .(((((((((((((.......)))))((..((((((.......)))))).))..............)))))))) ( -31.10, z-score = -3.31, R) >droPer1.super_25 195702 74 - 1448063 CGGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCACUAGACCAACGAGCCU .(((((((((((((.......)))))((..((((((.......)))))).))..............)))))))) ( -31.10, z-score = -3.31, R) >droWil1.scaffold_180764 989789 74 + 3949147 UGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCA ..((((((((((((.......)))))......((((.......))))...................))))))). ( -27.80, z-score = -2.52, R) >droVir3.scaffold_12970 6178752 74 + 11907090 UGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCA ..((((((((((((.......)))))......((((.......))))...................))))))). ( -27.80, z-score = -2.52, R) >droGri2.scaffold_15081 1374012 74 + 4274704 UGGGCUCGUCCGGGAGUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUGCCACUAGACCAACGAGCCA ..((((((((((((.......)))))((..((((((.......)))))).))..............))))))). ( -30.90, z-score = -2.92, R) >anoGam1.chrU 1854101 74 - 59568033 UGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCUAAGCGAAAAUCAUACCCCUAGACCAUUGAGCCG ..((((((((((((.......)))))......((((.......))))...................))))))). ( -25.10, z-score = -3.15, R) >consensus UGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAUACCCCUAGACCAACGAGCCA ..((((((((((((.......)))))......((((.......))))...................))))))). (-26.11 = -25.61 + -0.50)
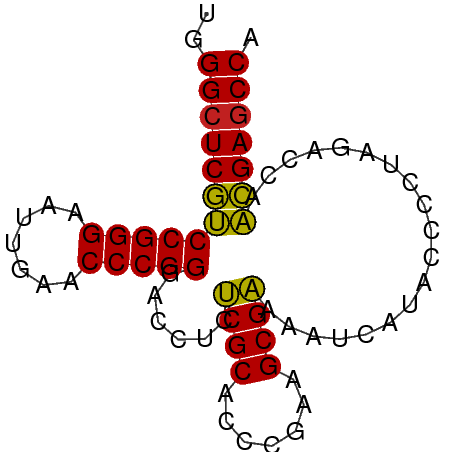
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:21 2011