
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,716,604 – 3,716,748 |
| Length | 144 |
| Max. P | 0.938357 |

| Location | 3,716,604 – 3,716,748 |
|---|---|
| Length | 144 |
| Sequences | 7 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 75.45 |
| Shannon entropy | 0.48008 |
| G+C content | 0.43971 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -21.74 |
| Energy contribution | -24.49 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
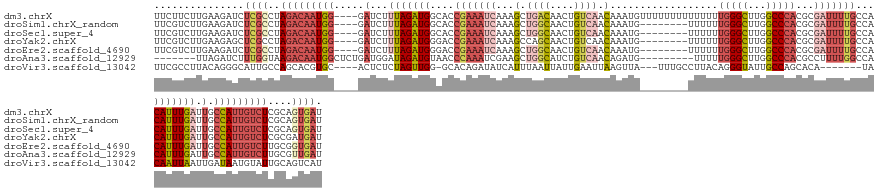
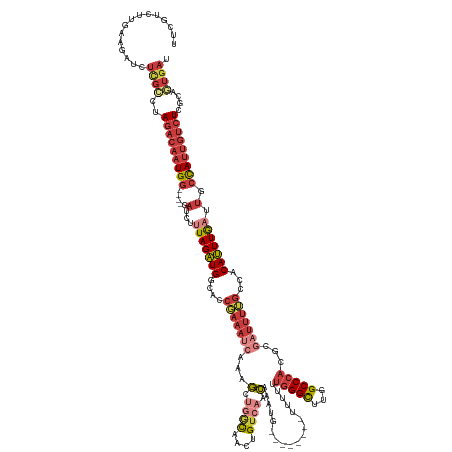
>dm3.chrX 3716604 144 - 22422827 UUCUUCUUGAAGAUCUCGCCUAGACAAUGG----GAUCUUUAGAUGGCACCGAAAUCAAAGCUGACAACUGUCAACAAAUGUUUUUUUUUUUUUUGGGCUUGGCCCACGCGAUUUUGCCACAUUUGAUUGCCAUUGUCUCGCAGUGAU .((((....))))..((((..(((((((((----.(...(((((((....(((((((...(.((((....)))).)..................(((((...)))))...)))))))...))))))).).)))))))))....)))). ( -40.10, z-score = -1.94, R) >droSim1.chrX_random 1508253 136 - 5698898 UUCGUCUUGAAGAUCUCGCCUAGACAAUGG----GAUCUUUAGAUGGCACCGAAAUCAAAGCUGGCAACUGUCAACAAAUG--------UUUUUUGGGCUUGGCCCACGCGAUUUUGCCACAUUUGAUUGCCAUUGUCUCGCAGUGAU .((((((.((((((((((.........)))----)))))))))))))......(((((((..((((((..((((((....)--------))...(((((...)))))...))).))))))..)))))))..(((((.....))))).. ( -41.90, z-score = -1.75, R) >droSec1.super_4 3183093 136 + 6179234 UUCGUCUUGAAGAUCUCGCCUAGACAAUGG----GAUCUUUAGAUGGCACCGAAAUCAAAGCUGGCAACUGUCAACAAAUG--------UUUUUUGGGCUUGGCCCACGCGAUUUUGCCACAUUUGAUUGCCAUUGUCUCGCAGUGAU .((((((.((((((((((.........)))----)))))))))))))......(((((((..((((((..((((((....)--------))...(((((...)))))...))).))))))..)))))))..(((((.....))))).. ( -41.90, z-score = -1.75, R) >droYak2.chrX 17070020 136 + 21770863 UUCGUCUUGAAGAGCUCGCCUAGACAAUGG----GAUCUUUAGAUGGGACCGAAAUCAAAGCCAGCAACUGUCAACAAAUG--------UUUUUUGGGCUUGGCCCACGCGAUUUUGCCACAUUUGAUUGCCAUUGUCUCGCGAUGAU .(((((..((.....))((..(((((((((----.(...(((((((((..(((((((......((((..((....))..))--------))...(((((...)))))...))))))))).))))))).).))))))))).))))))). ( -41.00, z-score = -1.44, R) >droEre2.scaffold_4690 1099721 136 - 18748788 UUCGUCUUGAAGAUCUCGCCUAGACAAUGG----GAUCUUUAGAUGGGACCGAAAUCAAAGCUGGCAACUGUCAACAAAUG--------UUUUUUGGGCUUGGCCCACGCGAUUUUGCCACAUUUGAUUGCCAUUGUCUUGCGGUGAU ...............(((((.(((((((((----.(...(((((((((..(((((((...(.((((....)))).).....--------.....(((((...)))))...))))))))).))))))).).)))))))))...))))). ( -44.10, z-score = -2.16, R) >droAna3.scaffold_12929 278292 132 - 3277472 -------UUAGAUCUUUGGUAAGACAAUGGCUCUGAUGGAUAGAUGUAACCCAAAUCGAAGCUGGCAUCUGUCAACAGAUG---------UUUUUGGGCUUGGCCCACGCCUUUUGGCCACAUUUGAUUGCCAUUGUCUUGCGUUGAU -------...........(((((((((((((.......(((((((((....(........)...)))))))))..((((((---------(...(((((...))))).(((....))).)))))))...)))))))))))))...... ( -49.70, z-score = -4.15, R) >droVir3.scaffold_13042 3398356 133 - 5191987 UUCGCCUUACAGGGCAUUGCCAGCACGUGC----ACUCUCUAGUUGG-GCACAGAUAUCAUUUAAUUAUUGAAUUAAGUUA---UUUGCCUUACAGGGUAUUGCCAGCACA-------UACAAUUAAUUGAUAAUGUAUUGCAGUCAU ............((((.((((.((....))----........((.((-(((..((((..((((((((....))))))))))---))))))).))..)))).)))).(((.(-------((((.(((.....)))))))))))...... ( -31.60, z-score = -0.79, R) >consensus UUCGUCUUGAAGAUCUCGCCUAGACAAUGG____GAUCUUUAGAUGGCACCGAAAUCAAAGCUGGCAACUGUCAACAAAUG________UUUUUUGGGCUUGGCCCACGCGAUUUUGCCACAUUUGAUUGCCAUUGUCUCGCAGUGAU ...............((((..(((((((((....((((...(((((....(((((((...(.((((....)))).)..................(((((...)))))...)))))))...))))))))).)))))))))....)))). (-21.74 = -24.49 + 2.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:19 2011