
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,713,725 – 3,713,812 |
| Length | 87 |
| Max. P | 0.998214 |

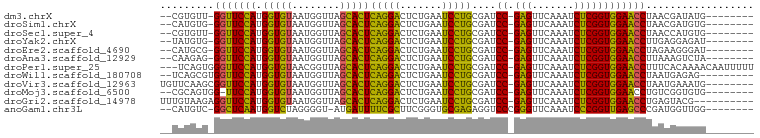
| Location | 3,713,725 – 3,713,812 |
|---|---|
| Length | 87 |
| Sequences | 12 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Shannon entropy | 0.42250 |
| G+C content | 0.49378 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -22.93 |
| Energy contribution | -22.74 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.965025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
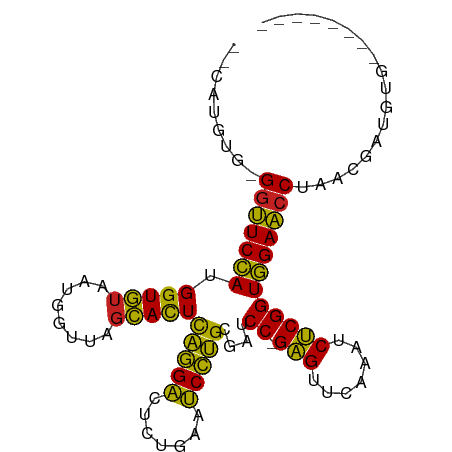
>dm3.chrX 3713725 87 + 22422827 --CGUGUU-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUAACGAUAUG-------- --(((...-(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))..))).....-------- ( -28.70, z-score = -2.47, R) >droSim1.chrX 2720931 87 + 17042790 --CAUGUG-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUAACGAUGUG-------- --(((.((-(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))))...)))..-------- ( -28.10, z-score = -1.81, R) >droSec1.super_4 3180317 87 - 6179234 --CGUGUU-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUAACCAUGUG-------- --((((..-(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))....))))..-------- ( -27.40, z-score = -1.91, R) >droYak2.chrX 17067195 87 - 21770863 --UAUGUG-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUUGAGGAGAU-------- --.....(-(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))).........-------- ( -26.10, z-score = -0.68, R) >droEre2.scaffold_4690 1096962 87 + 18748788 --CAUGCG-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUAGAAGGGAU-------- --.....(-(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))).........-------- ( -26.80, z-score = -0.75, R) >droAna3.scaffold_12929 275778 87 + 3277472 --CAAGAG-GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUUAAAGUCUA-------- --....((-(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))))........-------- ( -27.50, z-score = -1.91, R) >droPer1.super_25 186975 95 + 1448063 ---UCAGUGGGUUCCAUGGUGUAACGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUUUCACAAAACAAUUUUU ---...((((((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))...)))............ ( -27.20, z-score = -1.64, R) >droWil1.scaffold_180708 11651040 87 - 12563649 --UCAGCGUGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUAAUGAGAG--------- --(((....(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))...)))...--------- ( -27.40, z-score = -1.55, R) >droVir3.scaffold_12963 7605558 90 - 20206255 UGUUCAAGCGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUAAUGAAAUG-------- ..((((...(((((((.(((((........)))))(((((.......)))))....((-(((.......))))))))))))...))))...-------- ( -27.40, z-score = -2.14, R) >droMoj3.scaffold_6500 3021355 87 - 32352404 --CGCAGUGG-UUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUGUCGGUGUG-------- --(((...((-(((((.(((((........)))))(((((.......)))))....((-(((.......)))))))))))).....)))..-------- ( -27.20, z-score = -1.02, R) >droGri2.scaffold_14978 1057491 88 + 1124632 UUUGUAAGAGGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC-GAGUUCAAAUCUCGGUGGAACCUGAGUACG---------- ...(((..((((((((.(((((........)))))(((((.......)))))....((-(((.......)))))))))))))...))).---------- ( -28.90, z-score = -2.19, R) >anoGam1.chr3L 28727330 87 - 41284009 --CAUGUC-GGCUCAAUGGUCUAGGGGU-AUGAUUUUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGAUGGUUGG-------- --...(((-((((((((.......(((.-....(((((((.......)))))))..)))(((.......))).)))))).)))))......-------- ( -31.10, z-score = -0.77, R) >consensus __CAUGUG_GGUUCCAUGGUGUAAUGGUUAGCACUCAGGACUCUGAAUCCUGCGAUCC_GAGUUCAAAUCUCGGUGGAACCUAACGAUGUG________ .........(((((((((((((........)))))..((((((.(.(((....))).).))))))........)))))))).................. (-22.93 = -22.74 + -0.18)
| Location | 3,713,725 – 3,713,812 |
|---|---|
| Length | 87 |
| Sequences | 12 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Shannon entropy | 0.42250 |
| G+C content | 0.49378 |
| Mean single sequence MFE | -27.11 |
| Consensus MFE | -21.32 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3713725 87 - 22422827 --------CAUAUCGUUAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-AACACG-- --------......(((.((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))-)))...-- ( -28.40, z-score = -3.77, R) >droSim1.chrX 2720931 87 - 17042790 --------CACAUCGUUAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-CACAUG-- --------..........((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))-......-- ( -26.00, z-score = -2.63, R) >droSec1.super_4 3180317 87 + 6179234 --------CACAUGGUUAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-AACACG-- --------......(((.((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))-)))...-- ( -28.10, z-score = -3.19, R) >droYak2.chrX 17067195 87 + 21770863 --------AUCUCCUCAAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-CACAUA-- --------..........((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))-......-- ( -26.00, z-score = -2.56, R) >droEre2.scaffold_4690 1096962 87 - 18748788 --------AUCCCUUCUAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-CGCAUG-- --------..........((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))-......-- ( -26.00, z-score = -2.39, R) >droAna3.scaffold_12929 275778 87 - 3277472 --------UAGACUUUAAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC-CUCUUG-- --------..........((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))-......-- ( -26.00, z-score = -2.61, R) >droPer1.super_25 186975 95 - 1448063 AAAAAUUGUUUUGUGAAAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCGUUACACCAUGGAACCCACUGA--- ............(((...((((((((((((.......)))-)).....((.((((((....)))))).)).............))))))))))...--- ( -27.30, z-score = -1.86, R) >droWil1.scaffold_180708 11651040 87 + 12563649 ---------CUCUCAUUAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCACGCUGA-- ---------.........((((((((((((.......)))-)).....((.((((((....)))))).)).............))))))).......-- ( -26.90, z-score = -2.37, R) >droVir3.scaffold_12963 7605558 90 + 20206255 --------CAUUUCAUUAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCGCUUGAACA --------...((((...((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))))...)))).. ( -29.70, z-score = -3.80, R) >droMoj3.scaffold_6500 3021355 87 + 32352404 --------CACACCGACAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAA-CCACUGCG-- --------..........((((((((((((.......)))-)).....((.((((((....)))))).)).............)))))-))......-- ( -26.90, z-score = -2.82, R) >droGri2.scaffold_14978 1057491 88 - 1124632 ----------CGUACUCAGGUUCCACCGAGAUUUGAACUC-GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACCUCUUACAAA ----------.(((...(((((((((((((.......)))-)).....((.((((((....)))))).)).............))))))))..)))... ( -28.90, z-score = -3.44, R) >anoGam1.chr3L 28727330 87 + 41284009 --------CCAACCAUCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCGAAGCGAAAAUCAU-ACCCCUAGACCAUUGAGCC-GACAUG-- --------..........((((((((((((.......)))))......((((.......)))).......-............)))))))-......-- ( -25.10, z-score = -2.27, R) >consensus ________CACAUCGUUAGGUUCCACCGAGAUUUGAACUC_GGAUCGCAGGAUUCAGAGUCCUGAGUGCUAACCAUUACACCAUGGAACC_CACAUG__ ..................((((((((((((.......))).))....((((((.....))))))...................)))))))......... (-21.32 = -20.66 + -0.67)

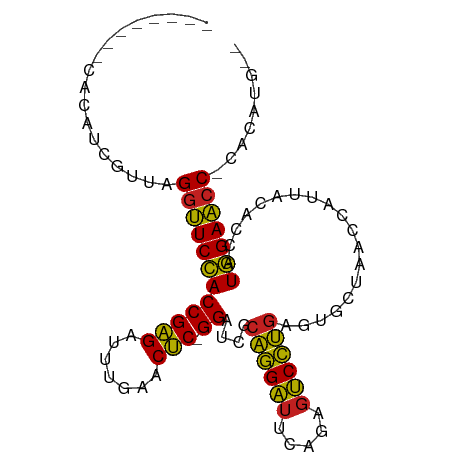
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:18 2011