| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,706,861 – 3,706,977 |
| Length | 116 |
| Max. P | 0.910991 |

| Location | 3,706,861 – 3,706,977 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.45 |
| Shannon entropy | 0.63357 |
| G+C content | 0.42208 |
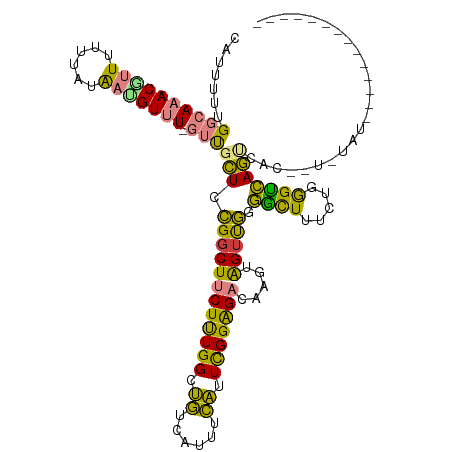
| Mean single sequence MFE | -26.57 |
| Consensus MFE | -14.01 |
| Energy contribution | -11.56 |
| Covariance contribution | -2.45 |
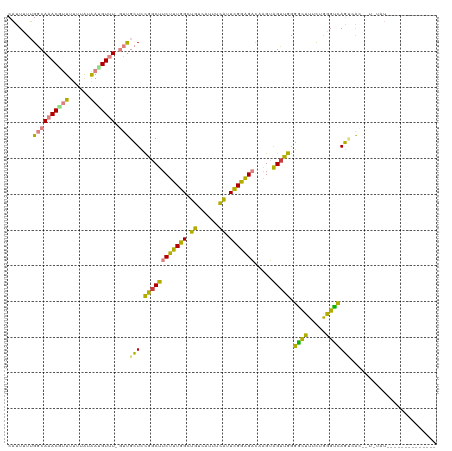
| Combinations/Pair | 1.61 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.910991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
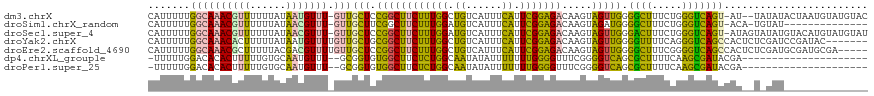
>dm3.chrX 3706861 116 + 22422827 CAUUUUUGGCAAACGUUUUUUAUAAUGUUU-GUUGCUCCGGCUUCUUUGGCUGUCAUUUCAUUCGGAGACAAGUAGUUGGGGCUUUCUGGGUCAGU-AU--UAUAUACUAAUGUAUGUAC .......((((((((((......)))))))-)))(((((((((((((((..((......))..)))))).....))))))))).............-..--(((((((....))))))). ( -32.30, z-score = -3.36, R) >droSim1.chrX_random 1499364 103 + 5698898 CAUUUUUGGCAAACGUUUUUUAUAACGUUU-GUUGCUUCGGCUUCUUUGGAUGUCAUUUCAUUCGGAGACAAGUAGAUGGGGCUUUCUGGGUCAGU-ACA-UGUAU-------------- .......((((((((((......)))))))-)))((....)).((((((((((......))))))))))...(((.(((.((((.....))))...-.))-).)))-------------- ( -30.30, z-score = -3.70, R) >droSec1.super_4 3173561 118 - 6179234 CAUUUUUGGCAAACGUUUUUUAUAACGUUU-GUUGCUCCGGCUUCUUUGGAUGUCAUUUCAUUCGGAGACAAGUAGUUGGGACUUUCUGGGUCAGU-AUAGUAUAUGUACAUGUAUGUAU .......((((((((((......)))))))-)))...((((((((((((((((......)))))))))).....))))))((((.....)))).((-(((.....))))).......... ( -33.10, z-score = -3.15, R) >droYak2.chrX 17058951 113 - 21770863 CAUUUUUGGCAAACACUUUUUAUAAUGUUUUGUUGCUGCGGCUUCUUUGGCUGUCAUUUCAUUCGGAGACAAGUAGUUGGGGUUUUCAGGGUCAGCCACUCUCGAUCCGAUAC------- .......((((((.((..........))))))))(((((....((((((..((......))..))))))...)))))..(((((...(((((.....))))).))))).....------- ( -21.00, z-score = 1.25, R) >droEre2.scaffold_4690 1089896 115 + 18748788 CAUUUUUGGCAAACGCUUUUUACGACGUUUUGUUGCUCCGGCUUCUUUGGCUGUCAUUUCAUUCGGAGACAAGUAGUUGGGGCUUUCGGGGUCAGCCACUCUCGAUGCGAUGCGA----- ........(((..(((..................(((((((((((((((..((......))..)))))).....)))))))))..((((((........)))))).))).)))..----- ( -30.10, z-score = 0.32, R) >dp4.chrXL_group1e 6843073 96 + 12523060 -UUUUUGGACACACUUUUUGUGCAAUGUUU--GCGGUGUGGCUUCUCUGGCAAUAUAUUUUUUUGGGGUUUCGGGGUCAGCGCUUUUCAAGCGAUACGA--------------------- -...(((.(((.......))).)))(((((--(.(((((((((((...(((..((........))..)))..))))))).))))...))))))......--------------------- ( -19.60, z-score = 0.50, R) >droPer1.super_25 178691 96 + 1448063 -UUUUUGGACACACUUUUUGUGCAAUGUUU--GCGGUGUGGCUUCUCUGGCAAUAUAUUUUUUUGGGGUUUCGGGGUCAGCGCUUUUCAAGCGAUACGA--------------------- -...(((.(((.......))).)))(((((--(.(((((((((((...(((..((........))..)))..))))))).))))...))))))......--------------------- ( -19.60, z-score = 0.50, R) >consensus CAUUUUUGGCAAACGUUUUUUAUAAUGUUU_GUUGCUCCGGCUUCUUUGGCUGUCAUUUCAUUCGGAGACAAGUAGUUGGGGCUUUCUGGGUCAGUCAC__U_UAU______________ .......((((((((((......)))))....))))).((((((((((((.((......)).))))))).....))))).((((.....))))........................... (-14.01 = -11.56 + -2.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:16 2011