| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,678,502 – 3,678,595 |
| Length | 93 |
| Max. P | 0.639338 |

| Location | 3,678,502 – 3,678,595 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 62.09 |
| Shannon entropy | 0.81883 |
| G+C content | 0.46121 |
| Mean single sequence MFE | -16.72 |
| Consensus MFE | -8.51 |
| Energy contribution | -8.00 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.08 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.639338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
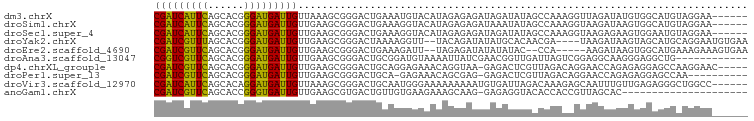
>dm3.chrX 3678502 93 - 22422827 CGAUCAUUCAGCACGGGAUGAUUGUUAAAGCGGGACUGAAAUGUACAUAGAGAGAUAGAUAUAGCCAAAGGUUAGAUAUGUGGCAUGUAGGAA------ (((((((((......)))))))))...........((...(((((((((............(((((...)))))..))))).))))...))..------ ( -17.84, z-score = -1.01, R) >droSim1.chrX 2699543 93 - 17042790 CGAUCAUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGAAAGGUACAUAGAGAGAUAAAUAUAGCCAAAGGUAAGAUAAGUGGCAUGUAGGAA------ (((((((((......))))))))).......(..(((....))).).............((((((((...((...))...)))).))))....------ ( -16.50, z-score = -1.16, R) >droSec1.super_4 3154857 93 + 6179234 CGAUCAUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGAAAGGUACAUAGAGAGAUAGAUAUAGCCAAAGGUAAGAGAAGUGGAAUGUAGGAA------ (((((((((......))))))))).....(((..(((....))).).................(((...))).......))............------ ( -12.00, z-score = 0.24, R) >droYak2.chrX 17037716 93 + 21770863 CGAUCGUUUAGCACGGGAUGAUUGUUGAAGCGGGACUAAAAGGUU--UACAGAUAUAUGCACAACGA----UAAGAUAAGUAGCAUGCAGGAAUGUGAA (((((((((......))))))))).....((.(((((....))))--)........((((((.....----........)).))))))........... ( -12.82, z-score = 1.02, R) >droEre2.scaffold_4690 1068798 90 - 18748788 CGAUCGUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGAAAGAUU--UAGAGAUAUAUAUAC--CCA-----AAGAUAAGUGGCAUGAAAGAAAGUGAA (((((((((......))))))))).......(((........(((--....))).......)--)).-----........................... ( -11.36, z-score = 0.92, R) >droAna3.scaffold_13047 139576 87 - 1816235 CGGUCGUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGCGGAUGUAAAAUUAUCGAACGGUUGAUUAGUCGGAGGCAAGGGAGGCUG------------ (((((..(((((.((.(((((((......(((....)))........)))))))...)))))))...((.....))......)))))------------ ( -19.24, z-score = 0.35, R) >dp4.chrXL_group1e 4032707 93 + 12523060 CGAUCGUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGCAGGAGAAACAGGUAA-GAGACUCGUUAGACAGGAACCAGAGAGGAGCCAAGGAAC----- .((((((((......))))))))(((..((((((.((((..(.....)..))..-.)).)))))).))).((..((.....))..)).......----- ( -18.80, z-score = 0.01, R) >droPer1.super_13 83093 87 - 2293547 CGAUCGUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGCA-GAGAAACAGCGAG-GAGACUCGUUAGACAGGAACCAGAGAGGAGCCAA---------- (((((((((......))))))))).....(((....))).-.......((((((-....)))))).....((..((.....))..))..---------- ( -25.20, z-score = -2.05, R) >droVir3.scaffold_12970 6994007 93 - 11907090 CGAUCAUUCAGCACAGGAUGAUUGUUAAAGCGGGACUGCAAUGGGAAAAAAAAAUGUGAUUAGACAAAGAGCAAUUUGUUGAGAGGGCUGGCC------ (((((((((......))))))))).....((.((.((.((((..(.........(((......)))........)..))))...)).)).)).------ ( -17.53, z-score = -0.16, R) >anoGam1.chrX 614033 77 - 22145176 CGAUCGUUCAGCACCGGGUGAUUGUUGAAGCGUGACUGUUGUGAAGAAAGCAAG-GAGAGGUACACCACCGUUAGCAC--------------------- ((((((..(......)..)))))).....((.((((((((........))))..-....(((.....)))))))))..--------------------- ( -15.90, z-score = 1.08, R) >consensus CGAUCGUUCAGCACGGGAUGAUUGUUGAAGCGGGACUGAAAGGUAAAUAGAGAGAUAGAUAUAACCAAAAGUAAGAUAAGUGGAAUGAAGGAA______ (((((((((......)))))))))........................................................................... ( -8.51 = -8.00 + -0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:13 2011