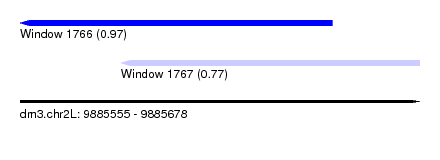
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 9,885,555 – 9,885,678 |
| Length | 123 |
| Max. P | 0.971538 |

| Location | 9,885,555 – 9,885,651 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.50 |
| Shannon entropy | 0.50633 |
| G+C content | 0.42429 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.36 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
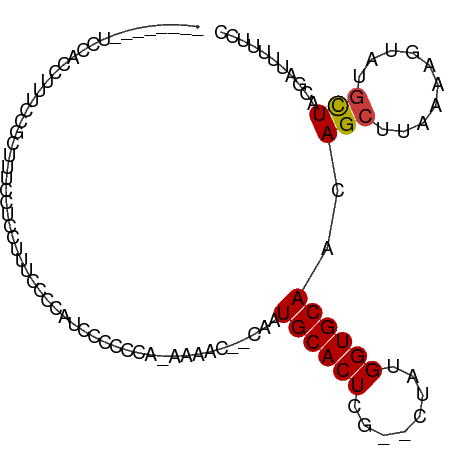
>dm3.chr2L 9885555 96 - 23011544 --------UUUGCCCACAAAAAC--CAAUGCACUCG--CUACGGUGCAACAGCUUAAAAGUAUGCUACGA-UUUUUCCCACUCAAUA---UUUUCGUGUAAUUGUUGCGCCU --------...............--....((....)--)...((((((((((((....))).(((.((((-................---...)))))))..))))))))). ( -18.91, z-score = -1.12, R) >droPer1.super_1 8525520 106 + 10282868 CUUUUUUCAUCCCAUUUAAAUCCCUCGCUGCACUCGCUCUCUGGUGCAACAGCUUAAAGG-AUGCUGCGA-UUUUUCUCACUCAACA----UUUCGUGUAAUUGUUGCGCCU ..........................((((((((........)))))...)))....(((-.(((.((((-((.....(((......----....))).)))))).)))))) ( -18.20, z-score = -0.29, R) >dp4.chr4_group3 11409048 106 + 11692001 CUUUUUUCAUCCCAUUUAAAUCCCUCGCUGCACUCGCUCUCUGGUGCAACAGCUUAAAGG-AUGCUGCGA-UUUUUCUCACUCAACA----UUUCGUGUAAUUGUUGCGCCU ..........................((((((((........)))))...)))....(((-.(((.((((-((.....(((......----....))).)))))).)))))) ( -18.20, z-score = -0.29, R) >droAna3.scaffold_12943 1330108 100 - 5039921 --------CUUUCGCCCAAAAAC--CAAUGCACUCG--CUCUGGUGCAACAGCUUAAAAGUGUAUUACGACUUCUCCCCACUCGACUCGAUUUUCGUGUAAUUGUUGCGCCU --------.....((......((--((..((....)--)..))))(((((((.....((((........))))......((.(((........))).))..))))))))).. ( -18.70, z-score = -0.96, R) >droEre2.scaffold_4929 10499199 93 - 26641161 ----------UUUCCCAACAAAC--CAAUGCACUCG--GUACGGUGCAACAGCUUAAAAGUAUGCUACGA-UUUUUCCCACUCAAUA----UUUCGUGUAAUUGUUGCGCCU ----------...........((--(.........)--))..((((((((((((....))).(((.((((-................----..)))))))..))))))))). ( -19.57, z-score = -1.59, R) >droYak2.chr2L 12576828 94 - 22324452 ---------UUUACCCACAAAAC--CAAUGCACUCG--CAUUGGUGCAACAGCUUAAAAGUAUGCUACGA-UUUUUCCCACUCAAUA----UUUCGUGUAAUUGUUGCGCCU ---------............((--((((((....)--)))))))(((((((((....))).(((.((((-................----..)))))))..)))))).... ( -22.97, z-score = -3.14, R) >droSec1.super_3 5331462 94 - 7220098 --------UUUCCCCACAAAAAC--CAAUGCACUCG--CUACGGUGCAACAGCUUAAAAGUAUGCUACGA--UUUUCCCACUCAAUA----UUUCGUGUAAUUGUUGCGCCU --------...............--....((....)--)...((((((((((((....))).(((.((((--...............----..)))))))..))))))))). ( -19.03, z-score = -2.07, R) >droSim1.chr2L 9669915 95 - 22036055 --------UUUCCCCACAAAAAC--CAAUGCACUCG--CUACGGUGCAACAGCUUAAAAGUAUGCUACGA-UUUUUCCCACUCAAUA----UUUCGUGUAAUUGUUGCGCCU --------...............--....((....)--)...((((((((((((....))).(((.((((-................----..)))))))..))))))))). ( -18.97, z-score = -2.00, R) >droVir3.scaffold_12963 18513239 80 + 20206255 ------------------------CUUUUGCAGUUUCACUUUGGUGCAACAGCUUAAAAGUAUGCUACGAUUUUUAUUCCCACA--------CAUACAUAAUUGUUGCGCUC ------------------------..................((((((((((.(((...(((((....((.......)).....--------))))).))))))))))))). ( -14.90, z-score = -1.04, R) >consensus _________UUCCCCACAAAAAC__CAAUGCACUCG__CUAUGGUGCAACAGCUUAAAAGUAUGCUACGA_UUUUUCCCACUCAAUA____UUUCGUGUAAUUGUUGCGCCU ..........................................((((((((((((....))).(((.((((.......................)))))))..))))))))). (-14.39 = -14.36 + -0.03)
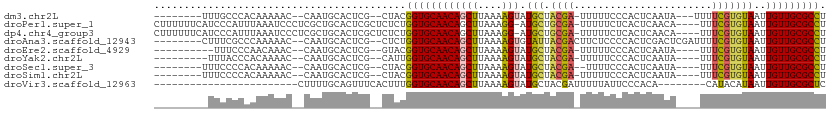
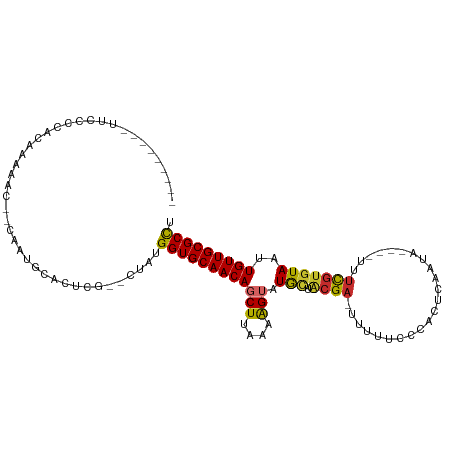
| Location | 9,885,586 – 9,885,678 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 66.97 |
| Shannon entropy | 0.62569 |
| G+C content | 0.46753 |
| Mean single sequence MFE | -13.24 |
| Consensus MFE | -7.38 |
| Energy contribution | -7.40 |
| Covariance contribution | 0.02 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 9885586 92 - 23011544 --------UCCAACUUUCCGCUUUCCUCCCGAAAUUUUGCCCACA-AAAAC--CAAUGCACUCG--CUACGGUGCAACAGCUUAAAAGUAUGCUACGAUUUUUCC --------....(((((..(((............(((((....))-)))..--...((((((..--....))))))..)))...)))))................ ( -12.50, z-score = -0.80, R) >droPer1.super_1 8525550 103 + 10282868 -UUCCUCUUGCCUCUUGCCUCUUCCCUCCUUUUUUCAUCCCAUUUAAAUCCCUCGCUGCACUCGCUCUCUGGUGCAACAGCUUAAAGG-AUGCUGCGAUUUUUCU -...................................................((((.(((..(.....(((......)))......).-.))).))))....... ( -13.60, z-score = 0.10, R) >dp4.chr4_group3 11409078 103 + 11692001 -UUCCUCUUGCCUCUUGCCUCUUCCCUCCUUUUUUCAUCCCAUUUAAAUCCCUCGCUGCACUCGCUCUCUGGUGCAACAGCUUAAAGG-AUGCUGCGAUUUUUCU -...................................................((((.(((..(.....(((......)))......).-.))).))))....... ( -13.60, z-score = 0.10, R) >droAna3.scaffold_12943 1330143 100 - 5039921 GUUCCCGUUUCUGGCUUCGUCCUUUCGCCUUUCGCCUUUCGCCCA-AAAAC--CAAUGCACUCG--CUCUGGUGCAACAGCUUAAAAGUGUAUUACGACUUCUCC .....(((....(((...........)))....((((((.((...-...((--((..((....)--)..))))......))...)))).))...)))........ ( -13.90, z-score = 0.52, R) >droEre2.scaffold_4929 10499229 84 - 26641161 ----------------UCCACUUUUCUGCUUUCCCCAUUUCCCAA-CAAAC--CAAUGCACUCG--GUACGGUGCAACAGCUUAAAAGUAUGCUACGAUUUUUCC ----------------...((((((..(((.......(((.....-.))).--...((((((..--....))))))..)))..))))))................ ( -11.40, z-score = -0.58, R) >droYak2.chr2L 12576858 93 - 22324452 --------UCCACUUUUCCGCUUCUCCGCUAUCCCCAUUUACCCACAAAAC--CAAUGCACUCG--CAUUGGUGCAACAGCUUAAAAGUAUGCUACGAUUUUUCC --------...((((((..(((.....((........(((......)))((--((((((....)--)))))))))...)))..))))))................ ( -16.00, z-score = -2.94, R) >droSec1.super_3 5331492 91 - 7220098 --------UCCACUUUUCCGCUUUCCCCUCCCCGAUUUCCCCACA-AAAAC--CAAUGCACUCG--CUACGGUGCAACAGCUUAAAAGUAUGCUACGAUUUUCC- --------...((((((..(((.((........))..........-.....--...((((((..--....))))))..)))..))))))...............- ( -11.70, z-score = -1.49, R) >consensus ________UCCACCUUUCCGCUUUCCUCCUUUCCCCAUCCCCCCA_AAAAC__CAAUGCACUCG__CUAUGGUGCAACAGCUUAAAAGUAUGCUACGAUUUUUCC ........................................................((((((........))))))..(((..........)))........... ( -7.38 = -7.40 + 0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:29:13 2011