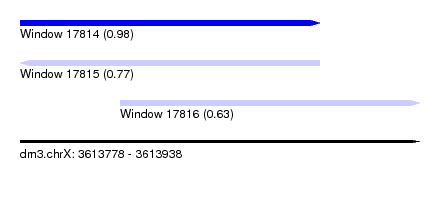
| Sequence ID | dm3.chrX |
|---|---|
| Location | 3,613,778 – 3,613,938 |
| Length | 160 |
| Max. P | 0.981818 |

| Location | 3,613,778 – 3,613,898 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.86 |
| Shannon entropy | 0.24920 |
| G+C content | 0.57356 |
| Mean single sequence MFE | -48.85 |
| Consensus MFE | -38.79 |
| Energy contribution | -38.48 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.981818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
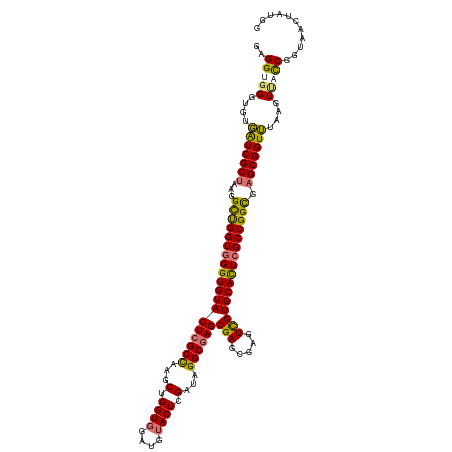
>dm3.chrX 3613778 120 + 22422827 GAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCUUUGCUGGUUGAUGUACUCGAUAGGCGAGCGAGCGAGUUGUGCACUCGCUGGCGAGCGGUUUAAGUAUCCGAUAACUAUGG .((.((((((.(((((((...((..((((((((((((((((((((.(((((.....))))).))))...))))))))...))))))))..)).)))))))....))))))....)).... ( -51.10, z-score = -3.71, R) >droSim1.chrX 2652958 120 + 17042790 GAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUAG ..(((((.(..(((((((...((..((((((((((((((...(((.((..((.......))..)).)))..))))))...))))))))..)).)))))))..).)))))........... ( -49.20, z-score = -2.04, R) >droSec1.super_4 3096042 120 - 6179234 GAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGAGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUGG ..(((((.(..(((((((...((..((((((((((.((((.....))))(((...((((.........))))...)))))))))))))..)).)))))))..).)))))........... ( -48.90, z-score = -2.37, R) >droEre2.scaffold_4690 1010188 116 + 18748788 GAGGUGGGUGUCGUCGCUAAGGUCGGUAGGUGUACUGGCUAAGCUGGUGCAUGUACUCGGUGGGCGAGCGA----UUCGUGCAUUCGCUGGUACGCGGUGUAUGUCCUCGCUAAAUGUGG ..((((((.(((((((((....((((((.((((((..((...))..)))))).))).)))..))))).)))----).(((((((.(((......))))))))))..))))))........ ( -46.20, z-score = -2.19, R) >consensus GAGGUGGGUGUGAUCGCUAAGGCUGGUGGGUGUACUCGCCAAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAACUAUGG ..(((((....(((((((...(((((((((((((((((((...(.((((....)))).)...))))))(((.....)))))))))))))))).)))))))....)))))........... (-38.79 = -38.48 + -0.31)

| Location | 3,613,778 – 3,613,898 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.86 |
| Shannon entropy | 0.24920 |
| G+C content | 0.57356 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -17.88 |
| Energy contribution | -21.50 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chrX 3613778 120 - 22422827 CCAUAGUUAUCGGAUACUUAAACCGCUCGCCAGCGAGUGCACAACUCGCUCGCUCGCCUAUCGAGUACAUCAACCAGCAAAGCGAGUACACCCACCAGCCUUAGCGAUCACACCCACCUC .....((.(((.............((((((.(((((((.....))))))).(((((.....)))))...............))))))..........((....))))).))......... ( -28.80, z-score = -2.95, R) >droSim1.chrX 2652958 120 - 17042790 CUAUAGCUACCGGUAACUUAAACCGCUCGCCAGCGAGUGCACGACUCGCUCGCUCGCCUAUCGAGUACAUCCACCAGCUUGGCGAGUACACCCACCAGCCUUAGCGAUCACACCCACCUC (((..(((...(((.......)))((((((((((((((.....))))))..(((((.....))))).............)))))))).........)))..)))................ ( -34.90, z-score = -3.03, R) >droSec1.super_4 3096042 120 + 6179234 CCAUAGCUACCGGUAACUUAAACCGCUCGCCAGCGAGUGCACGACUCUCUCGCUCGCCUAUCGAGUACAUCCACCAGCUUGGCGAGUACACCCACCAGCCUUAGCGAUCACACCCACCUC .....((((..(((.......)))((((((((((((((.....))))....(((((.....)))))..........)).))))))))..............))))............... ( -32.20, z-score = -2.83, R) >droEre2.scaffold_4690 1010188 116 - 18748788 CCACAUUUAGCGAGGACAUACACCGCGUACCAGCGAAUGCACGAA----UCGCUCGCCCACCGAGUACAUGCACCAGCUUAGCCAGUACACCUACCGACCUUAGCGACGACACCCACCUC ...........((((........(((......)))..........----(((.((((....((.(((..((.((..((...))..)).))..)))))......)))))))......)))) ( -20.30, z-score = -0.65, R) >consensus CCAUAGCUACCGGUAACUUAAACCGCUCGCCAGCGAGUGCACGACUCGCUCGCUCGCCUAUCGAGUACAUCCACCAGCUUAGCGAGUACACCCACCAGCCUUAGCGAUCACACCCACCUC .....((((..((((.....))))(((((((((((((((.......)))))(((((.....)))))..........)).))))))))..............))))............... (-17.88 = -21.50 + 3.62)

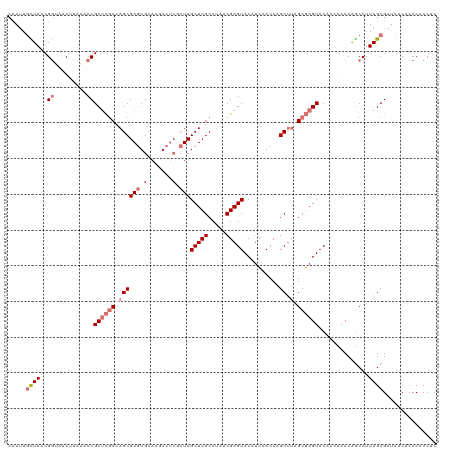
| Location | 3,613,818 – 3,613,938 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Shannon entropy | 0.30903 |
| G+C content | 0.54842 |
| Mean single sequence MFE | -46.42 |
| Consensus MFE | -31.77 |
| Energy contribution | -33.65 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.628923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
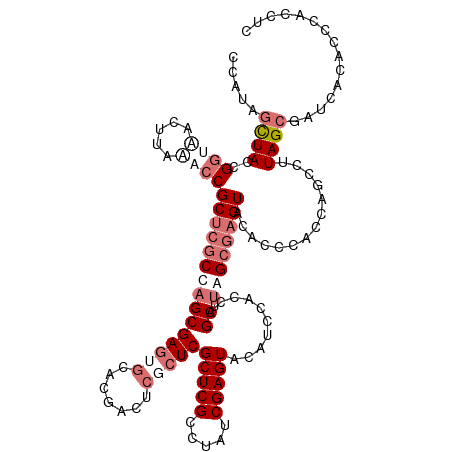
>dm3.chrX 3613818 120 + 22422827 UUGCUGGUUGAUGUACUCGAUAGGCGAGCGAGCGAGUUGUGCACUCGCUGGCGAGCGGUUUAAGUAUCCGAUAACUAUGGUUGGUGCACACGCUAAGUAACAAGUGACCGCUAAACUGCU (((((.(((((.....))))).)))))((.(((((((.....))))))).)).(((((((((.(((.(((((.......))))))))...((((........)))).....))))))))) ( -41.80, z-score = -1.22, R) >droSim1.chrX 2652998 120 + 17042790 AAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUAGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAACCGCU ..((..(((..(((((..(((..((......))..))))))))..)))..)).((((((((.(((((....)))))...((((((.((((.(........).)))))))))))))))))) ( -49.40, z-score = -2.06, R) >droSec1.super_4 3096082 120 - 6179234 AAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGAGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAGCUAUGGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAACCGCC ..((..(((..(((((......(((...(....).))))))))..)))..))..(((((((.(((((....)))))...((((((.((((.(........).))))))))))))))))). ( -46.50, z-score = -1.59, R) >droEre2.scaffold_4690 1010228 116 + 18748788 AAGCUGGUGCAUGUACUCGGUGGGCGAGCGAU----UCGUGCAUUCGCUGGUACGCGGUGUAUGUCCUCGCUAAAUGUGGUUUGUGCGCUCGCUGCAUUGCAUGUGAUUGCCAAACCGCG ..(((((((....))).)))).((((((.((.----.(((((((.(((......))))))))))))))))))....((((((((.(((.(((((((...))).)))).))))))))))). ( -48.00, z-score = -1.79, R) >consensus AAGCUGGUGGAUGUACUCGAUAGGCGAGCGAGCGAGUCGUGCACUCGCUGGCGAGCGGUUUAAGUUACCGGUAACUAUGGGUGGUGCACUCGCUAAGUAACAAGUGACCGCCAAACCGCU ..((..(((..((((((((.....))))(((.....)))))))..)))..)).((((((((.(((((....)))))...((((((.((((.(........).)))))))))))))))))) (-31.77 = -33.65 + 1.88)

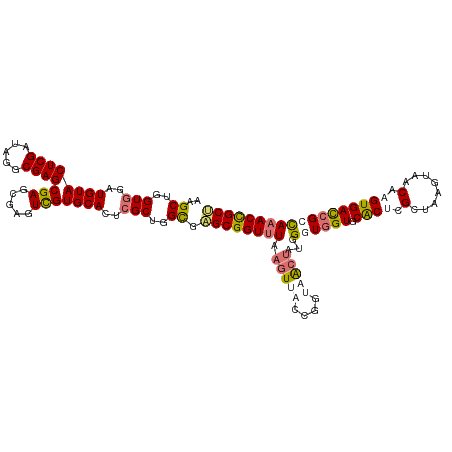
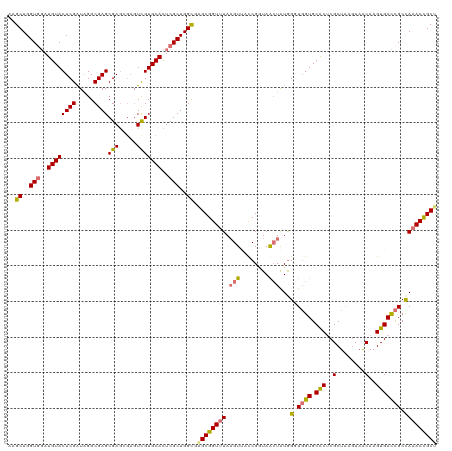
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 01:12:01 2011